You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_03893
You are here: Home > Sequence: MGYG000001436_03893
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
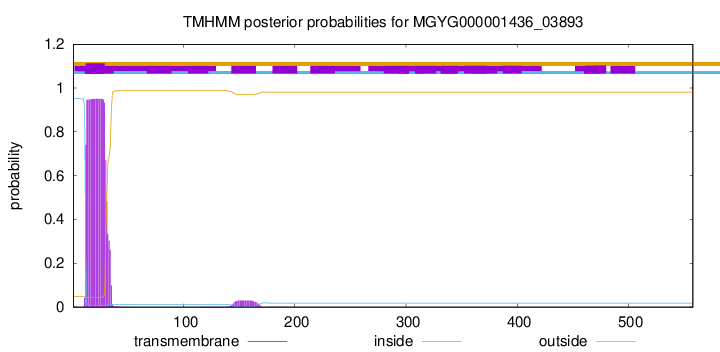
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_03893 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1070286; End: 1071962 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd12215 | ChiC_BD | 1.82e-15 | 46 | 89 | 1 | 42 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
| cd00063 | FN3 | 7.66e-13 | 106 | 190 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 3.10e-10 | 106 | 177 | 1 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| smart00495 | ChtBD3 | 4.42e-10 | 46 | 87 | 2 | 41 | Chitin-binding domain type 3. |
| cd00598 | GH18_chitinase-like | 1.14e-09 | 203 | 513 | 1 | 207 | The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV17644.1 | 0.0 | 1 | 558 | 1 | 560 |
| BAC76693.1 | 0.0 | 1 | 548 | 1 | 552 |
| AFI56073.1 | 0.0 | 1 | 548 | 1 | 552 |
| BBH85516.1 | 4.63e-284 | 18 | 558 | 20 | 554 |
| AFH63946.2 | 4.51e-246 | 109 | 558 | 193 | 642 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1K85_A | 2.34e-27 | 105 | 190 | 3 | 88 | ChainA, CHITINASE A1 [Niallia circulans] |
| 4TXG_A | 1.87e-10 | 26 | 109 | 33 | 113 | CrystalStructure of a Family GH18 Chitinase from Chromobacterium violaceum [Chromobacterium violaceum ATCC 12472] |
| 2D49_A | 1.87e-08 | 47 | 84 | 6 | 43 | ChainA, chitinase C [Streptomyces griseus] |
| 1WVU_A | 8.33e-07 | 47 | 84 | 6 | 43 | Crystalstructure of chitinase C from Streptomyces griseus HUT6037 [Streptomyces griseus],1WVU_B Crystal structure of chitinase C from Streptomyces griseus HUT6037 [Streptomyces griseus],2DBT_A Crystal structure of chitinase C from Streptomyces griseus HUT6037 [Streptomyces griseus],2DBT_B Crystal structure of chitinase C from Streptomyces griseus HUT6037 [Streptomyces griseus],2DBT_C Crystal structure of chitinase C from Streptomyces griseus HUT6037 [Streptomyces griseus] |
| 1WVV_A | 8.33e-07 | 47 | 84 | 6 | 43 | ChainA, chitinase C [Streptomyces griseus],1WVV_B Chain B, chitinase C [Streptomyces griseus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 3.57e-37 | 48 | 427 | 34 | 389 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| P20533 | 5.04e-25 | 102 | 197 | 556 | 651 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P50401 | 4.64e-17 | 98 | 196 | 471 | 572 | Exoglucanase A OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhA PE=1 SV=1 |
| P50899 | 5.23e-17 | 100 | 198 | 698 | 799 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
| P36909 | 1.77e-15 | 105 | 342 | 141 | 396 | Chitinase C OS=Streptomyces lividans OX=1916 GN=chiC PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000322 | 0.998951 | 0.000182 | 0.000215 | 0.000166 | 0.000146 |