You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_04023
Basic Information
help
| Species |
Paenibacillus_F sp000411255
|
| Lineage |
Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255
|
| CAZyme ID |
MGYG000001436_04023
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 793 |
|
80906.86 |
7.1157 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001436 |
6290841 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1209099;
End: 1211480
Strand: +
|
No EC number prediction in MGYG000001436_04023.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
11 |
261 |
6.5e-32 |
0.43148148148148147 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.18e-45 |
8 |
566 |
5 |
441 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
2.45e-33 |
66 |
224 |
1 |
159 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1928
|
PMT1 |
0.009 |
1 |
156 |
15 |
180 |
Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O34575
|
1.90e-202 |
5 |
760 |
4 |
675 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
|
P37483
|
7.43e-191 |
3 |
784 |
2 |
684 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.977814
|
0.008661
|
0.000756
|
0.000071
|
0.000039
|
0.012672
|
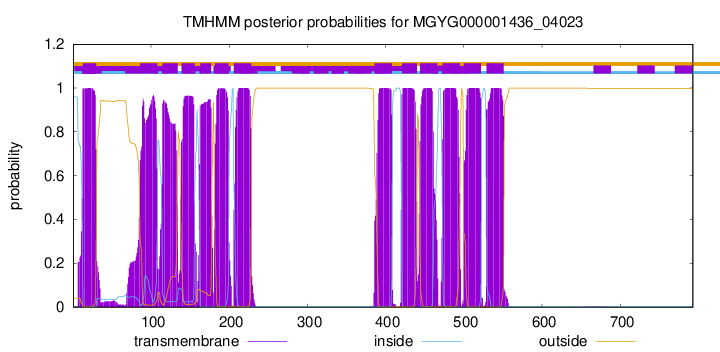
| start |
end |
| 13 |
30 |
| 86 |
108 |
| 115 |
134 |
| 139 |
156 |
| 163 |
177 |
| 182 |
199 |
| 206 |
228 |
| 386 |
408 |
| 421 |
440 |
| 444 |
466 |
| 473 |
495 |
| 500 |
522 |
| 529 |
551 |


