You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_04066
You are here: Home > Sequence: MGYG000001436_04066
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_04066 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1252271; End: 1254583 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3227 | LasB | 2.01e-161 | 1 | 526 | 1 | 507 | Zn-dependent metalloprotease [Posttranslational modification, protein turnover, chaperones]. |
| cd09597 | M4_TLP | 2.21e-123 | 257 | 525 | 1 | 278 | Peptidase M4 family including thermolysin, protealysin, aureolysin, and neutral protease. This peptidase M4 family includes several endopeptidases such as thermolysin (EC 3.4.24.27), aureolysin (the extracellular metalloproteinase from Staphylococcus aureus), neutral protease from Bacillus cereus, protealysin, and bacillolysin (EC 3.4.24.28). Typically, the M4 peptidases consist of a presequence (signal sequence), a propeptide sequence, and a peptidase unit. The presequence is cleaved off during export while the propeptide has inhibitory and chaperone functions and facilitates folding. The propeptide remains attached until the peptidase is secreted and can be safely activated. All peptidases in this family bind a single catalytic zinc ion which is tetrahedrally co-ordinated by three amino acid ligands and a water molecule that forms the nucleophile on activation during catalysis. The active site is found between two sub-domains; the N-terminal domain contains the HEXXH zinc-binding motif while the helical C-terminal domain, which is unique for the family, carries the third zinc ligand. These peptidases are secreted eubacterial endopeptidases from Gram-positive or Gram-negative sources that degrade extracellular proteins and peptides for bacterial nutrition. They are selectively inhibited by Steptomyces metalloproteinase inhibitor (SMPI) as well as by phosphoramidon from Streptomyces tanashiensis. A large number of these enzymes are implicated as key factors in the pathogenesis of various diseases, including gastritis, peptic ulcer, gastric carcinoma, cholera and several types of bacterial infections, and are therefore important drug targets. Some enzymes of the family can function at extremes of temperatures, while some function in organic solvents, thus rendering them novel targets for biotechnological applications. Thermolysin is widely used as a nonspecific protease to obtain fragments for peptide sequencing. It has also been used in production of the artificial sweetener aspartame. |
| pfam02868 | Peptidase_M4_C | 6.03e-78 | 368 | 525 | 1 | 167 | Thermolysin metallopeptidase, alpha-helical domain. |
| cd02699 | M4_M36 | 8.63e-72 | 257 | 524 | 1 | 313 | Peptidase M4 family (includes thermolysin, aureolysin, neutral protease and bacillolysin) and Peptidase M36 family (also known as fungalysin). This family includes the peptidases M4 as well as M36, both belonging to the Gluzincin family. The M4 peptidase family includes numerous zinc-dependent metallopeptidases that hydrolyze peptide bonds, such as thermolysin (EC 3.4.24.27), pseudolysin (the extracellullar elastase of Pseudomonas aeruginosa), aureolysin (the extracellular metalloproteinase from Staphylococcus aureus), neutral protease from Bacillus cereus, as well as bacillolysin (EC 3.4.24.28). The M36 family also known as fungalysin (elastinolytic metalloproteinase) family, includes endopeptidases from pathogenic fungi. Both M4 and M36 families have similar folds and contain the Zn-binding site and the active site HEXXH motif. The eukaryotic M36 and bacterial M4 families of metalloproteases also share a conserved domain in their propeptides called FTP (fungalysin/thermolysin propeptide). |
| pfam01447 | Peptidase_M4 | 8.17e-50 | 225 | 365 | 1 | 147 | Thermolysin metallopeptidase, catalytic domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV17803.1 | 0.0 | 1 | 770 | 1 | 770 |
| AIT70967.1 | 1.35e-85 | 526 | 766 | 452 | 693 |
| BCK52214.1 | 1.88e-85 | 526 | 765 | 452 | 692 |
| APO42946.1 | 2.07e-82 | 526 | 768 | 455 | 698 |
| BAA09831.1 | 2.07e-82 | 526 | 768 | 455 | 698 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4B52_A | 2.19e-145 | 223 | 525 | 1 | 303 | Crystalstructure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa [Paenibacillus polymyxa],4B52_B Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa [Paenibacillus polymyxa],4GER_A Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa [Paenibacillus polymyxa],4GER_B Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa [Paenibacillus polymyxa] |
| 5A3Y_A | 3.12e-135 | 39 | 525 | 42 | 547 | SADstructure of Thermolysin obtained by multi crystal data collection [Bacillus thermoproteolyticus] |
| 1NPC_A | 1.07e-118 | 225 | 525 | 8 | 316 | THESTRUCTURE OF NEUTRAL PROTEASE FROM BACILLUS CEREUS AT 0.2-NM RESOLUTION [Bacillus cereus] |
| 1ESP_A | 5.97e-118 | 225 | 525 | 8 | 316 | ChainA, NEUTRAL PROTEASE MUTANT E144S [Bacillus cereus] |
| 1KEI_A | 4.30e-114 | 225 | 525 | 8 | 315 | Thermolysin(substrate-free) [Bacillus thermoproteolyticus],1KJO_A Thermolysin complexed with Z-L-Threonine (benzyloxycarbonyl-L-Threonine) [Bacillus thermoproteolyticus],1KJP_A Thermolysin complexed with Z-L-Glutamic acid (benzyloxycarbonyl-L-Glutamic acid) [Bacillus thermoproteolyticus],1KKK_A Thermolysin complexed with Z-L-Aspartic Acid (benzyloxycarbonyl-L-Aspartic Acid) [Bacillus thermoproteolyticus],1KL6_A Thermolysin complexed with Z-L-Alanine (benzyloxycarbonyl-L-Alanine) [Bacillus thermoproteolyticus],1KR6_A Thermolysin complexed with Z-D-Glutamic acid (benzyloxycarbonyl-D-Glutamic acid) [Bacillus thermoproteolyticus],1KRO_A Thermolysin complexed with Z-D-Threonine (benzyloxycarbonyl-D-Threonine) [Bacillus thermoproteolyticus],1KS7_A Thermolysin complexed with Z-D-Aspartic acid (benzyloxycarbonyl-D-Aspartic acid) [Bacillus thermoproteolyticus],1KTO_A Thermolysin complexed with Z-D-Alanine (benzyloxycarbonyl-D-Alanine) [Bacillus thermoproteolyticus],1Y3G_E Crystal Structure of a Silanediol Protease Inhibitor Bound to Thermolysin [Bacillus thermoproteolyticus],2WHZ_A Dipeptide Inhibitors of Thermolysin [Bacillus thermoproteolyticus],2WI0_A Dipeptide Inhibitors of Thermolysin [Bacillus thermoproteolyticus],3DNZ_A Thermolysin by LB nanotemplate method before high X-Ray dose on ESRF ID14-2 beamline [Bacillus thermoproteolyticus],3DO0_A Thermolysin by classical hanging drop method after high X-Ray dose on esrf ID14-2 beamline [Bacillus thermoproteolyticus],3DO1_A Thermolysin by Classical hanging drop method before high X-Ray dose on ESRF ID14-2 beamline [Bacillus thermoproteolyticus],3DO2_A Thermolysin by LB nanotemplate method after high X-Ray dose on ESRF ID14-2 beamline [Bacillus thermoproteolyticus],3FB0_A Metal exchange in thermolysin [Bacillus thermoproteolyticus],3FBO_A Metal exchange in Thermolysin [Bacillus thermoproteolyticus],3FGD_A Drugscore FP: Thermoylsin in complex with fragment. [Bacillus thermoproteolyticus],3FLF_A Thermolysin inhibition [Bacillus thermoproteolyticus],3FV4_A Thermolysin inhibition [Bacillus thermoproteolyticus],3FVP_A Thermolysin inhibition [Bacillus thermoproteolyticus],3FXP_A Thermolysin inhibition [Bacillus thermoproteolyticus],3FXS_A Metal exchange in thermolysin [Bacillus thermoproteolyticus],3LS7_A Crystal structure of Thermolysin in complex with Xenon [Bacillus thermoproteolyticus],3MS3_A Crystal structure of Thermolysin in complex with Aniline [Bacillus thermoproteolyticus],3MSA_A Crystal structure of Thermolysin in complex with 3-Bromophenol [Bacillus thermoproteolyticus],3MSF_A Crystal structure of Thermolysin in complex with Urea [Bacillus thermoproteolyticus],3MSN_A Crystal structure of Thermolysin in complex with N-methylurea [Bacillus thermoproteolyticus],3N21_A Crystal structure of Thermolysin in complex with S-1,2-Propandiol [Bacillus thermoproteolyticus],3NN7_A Crystal structure of Thermolysin in complex with 2-bromoacetate [Bacillus thermoproteolyticus],3QGO_A Structure of Thermolysin in complex with L-Phenylalanine methylester [Bacillus thermoproteolyticus],3QH1_A Structure of Thermolysin in complex with N-benzyloxycarbonyl-L-aspartic acid [Bacillus thermoproteolyticus],3QH5_A Structure of Thermolysin in complex with N-Carbobenzyloxy-L-aspartic acid and L-Phenylalanine Methyl Ester [Bacillus thermoproteolyticus],3SSB_A Structure of Insect Metalloproteinase Inhibitor in Complex with Thermolysin [Bacillus thermoproteolyticus],3SSB_B Structure of Insect Metalloproteinase Inhibitor in Complex with Thermolysin [Bacillus thermoproteolyticus],3T73_A Thermolysin In Complex With UBTLN22 [Bacillus thermoproteolyticus],3T74_A Thermolysin In Complex With UBTLN27 [Bacillus thermoproteolyticus],3T87_A Thermolysin In Complex With UBTLN28 [Bacillus thermoproteolyticus],3T8C_A Thermolysin In Complex With UBTLN30 [Bacillus thermoproteolyticus],3T8D_A Thermolysin In Complex With UBTLN31 [Bacillus thermoproteolyticus],3T8F_A Thermolysin In Complex With UBTLN34 [Bacillus thermoproteolyticus],3T8G_A Thermolysin In Complex With UBTLN26 [Bacillus thermoproteolyticus],3T8H_A Thermolysin In Complex With UBTLN29 [Bacillus thermoproteolyticus],4D91_A Thermolysin In Complex With DMSO And Acetate [Bacillus thermoproteolyticus],4D9W_A Thermolysin In Complex With UBTLN32 [Bacillus thermoproteolyticus],4H57_A Thermolysin inhibition [Bacillus thermoproteolyticus],4MTW_E Thermolysin in complex with UBTLN36 [Bacillus thermoproteolyticus],4MWP_E Thermolysin in complex with UBTLN46 [Bacillus thermoproteolyticus],4MXJ_E Thermolysin in complex with UBTLN35 [Bacillus thermoproteolyticus],4MZN_E Thermolysin in complex with UBTLN59 [Bacillus thermoproteolyticus],4N4E_E Thermolysin in complex with UBTLN58 [Bacillus thermoproteolyticus],4N5P_E Thermolysin in complex with UBTLN20 [Bacillus thermoproteolyticus],4N66_E Thermolysin in complex with UBTLN37 [Bacillus thermoproteolyticus],4OI5_E Glycerol-free structure of thermolysin in complex with ubtln58 [Bacillus thermoproteolyticus],4OW3_A Thermolysin structure determined by free-electron laser [Bacillus thermoproteolyticus],4TNL_A 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL [Bacillus thermoproteolyticus],5DLH_A SFX structure of thermolysin [Geobacillus stearothermophilus],5DPE_E Thermolysin in complex with inhibitor. [Bacillus thermoproteolyticus],5DPF_E Thermolysin in complex with inhibitor. [Bacillus thermoproteolyticus],5FSJ_A Structure of thermolysin prepared by the 'soak-and-freeze' method under 45 bar of oxygen pressure [Bacillus thermoproteolyticus],5FSP_A Structure of thermolysin prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure [Bacillus thermoproteolyticus],5FSS_A Structure of thermolysin prepared by the 'soak-and-freeze' method under 40 bar of krypton pressure [Bacillus thermoproteolyticus],5JS3_E Thermolysin in complex with JC114. [Bacillus thermoproteolyticus],5JSS_E Thermolysin in complex with JC149. [Bacillus thermoproteolyticus],5JT9_E Thermolysin in complex with JC106. [Bacillus thermoproteolyticus],5JVI_E Thermolysin in complex with JC148. [Bacillus thermoproteolyticus],5JXN_E Thermolysin in complex with JC240. [Bacillus thermoproteolyticus],5K7T_A MicroED structure of thermolysin at 2.5 A resolution [Bacillus thermoproteolyticus],5L3U_E Thermolysin in complex with JC149 (MPD cryo protectant) [Bacillus thermoproteolyticus],5L41_E Thermolysin in complex with JC148 (MPD cryo protectant) [Bacillus thermoproteolyticus],5L8P_E Thermolysin in complex with JC114 (PEG400 cryo protectant) [Bacillus thermoproteolyticus],5LIF_E Thermolysin in complex with inhibitor [Bacillus thermoproteolyticus],5LVD_E Thermolysin in complex with inhibitor (JC67) [Bacillus thermoproteolyticus],5LWD_E Thermolysin in complex with inhibitor (JC96) [Bacillus thermoproteolyticus],5M5F_E Thermolysin in complex with inhibitor and krypton [Bacillus thermoproteolyticus],5M69_E Thermolysin in complex with inhibitor and xenon [Bacillus thermoproteolyticus],5M9W_A Experimental MAD phased structure of thermolysin in complex with inhibitor JC65. [Bacillus thermoproteolyticus],5MA7_E Structure of thermolysin in complex with inhibitor (JC306). [Bacillus thermoproteolyticus],5MNR_E Thermolysin in complex with inhibitor JC256 [Bacillus thermoproteolyticus],5N2T_E Thermolysin in complex with inhibitor JC287 [Bacillus thermoproteolyticus],5N2X_E Thermolysin in complex with inhibitor JC272 [Bacillus thermoproteolyticus],5N2Z_E Thermolysin in complex with inhibitor JC286 [Bacillus thermoproteolyticus],5N31_E Thermolysin in complex with inhibitor JC277 [Bacillus thermoproteolyticus],5N34_E Thermolysin in complex with inhibitor JC276 [Bacillus thermoproteolyticus],5N3V_E Thermolysin in complex with inhibitor JC292 [Bacillus thermoproteolyticus],5N3Y_E Thermolysin in complex with inhibitor JC267 [Bacillus thermoproteolyticus],5O8N_A Structure of thermolysin at room temperature via a method of acoustically induced rotation. [Bacillus thermoproteolyticus],5ONP_A Alzheimer's Amyloid-Beta Peptide Fragment 1-40 in Complex with Cd-substituted Thermolysin [Geobacillus stearothermophilus],5ONQ_A Alzheimer's Amyloid-Beta Peptide Fragment 29-40 in Complex with Cd-substituted Thermolysin [Geobacillus stearothermophilus],5ONR_A Alzheimer's Amyloid-Beta Peptide Fragment 1-40 in Complex with Thermolysin [Bacillus thermoproteolyticus],5T9I_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5T9K_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5T9Q_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAC_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAD_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAE_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAI_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAJ_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5TAK_A Conformational Sampling Differences across the Arrhenius Plot Biphasic Break Point at Ambient Temperature in the Enzyme Thermolysin [Bacillus thermoproteolyticus],5UN3_A Tetragonal thermolysin (295 K) in the presence of 50% xylose [Bacillus thermoproteolyticus],5UU7_A Tetragonal thermolysin (295 K) in the presence of 50% mpd [Bacillus thermoproteolyticus],5UU8_A Tetragonal thermolysin cryocooled to 100 K with 30% xylose as cryoprotectant [Bacillus thermoproteolyticus],5UU9_A Tetragonal thermolysin cryocooled to 100 K with 40% xylose as cryoprotectant [Bacillus thermoproteolyticus],5UUA_A Tetragonal thermolysin cryocooled to 100 K with 50% xylose as cryoprotectant [Bacillus thermoproteolyticus],5UUB_A Tetragonal thermolysin cryocooled to 100 K with 25% xylose/25% mpd as cryoprotectant [Bacillus thermoproteolyticus],5UUC_A Tetragonal thermolysin cryocooled to 100 K with 50% mpd as cryoprotectant [Bacillus thermoproteolyticus],5UUD_A Tetragonal thermolysin cryocooled to 100 K with 50% dmf as cryoprotectant [Bacillus thermoproteolyticus],5UUE_A Tetragonal thermolysin cryocooled to 100 K with 50% methanol as cryoprotectant [Bacillus thermoproteolyticus],5WR2_A Thermolysin, SFX liganded form with oil-based carrier [Geobacillus stearothermophilus],5WR3_A Thermolysin, SFX liganded form with water-based carrier [Geobacillus stearothermophilus],5WR4_A Thermolysin, SFX unliganded form with oil-based carrier [Geobacillus stearothermophilus],5WR5_A Thermolysin, liganded form with cryo condition 1 [Geobacillus stearothermophilus],5WR6_A Thermolysin, liganded form with cryo condition 2 [Geobacillus stearothermophilus],6D5N_A Hexagonal thermolysin (295) in the presence of 50% xylose [Bacillus thermoproteolyticus],6D5O_A Hexagonal thermolysin (295 K) in the presence of 50% DMF [Bacillus thermoproteolyticus],6D5P_A Hexagonal thermolysin cryocooled to 100 K with 20% xylose as cryoprotectant [Bacillus thermoproteolyticus],6D5Q_A Hexagonal thermolysin cryocooled to 100 K with 30% xylose as cryoprotectant [Bacillus thermoproteolyticus],6D5R_A Hexagonal thermolysin cryocooled to 100 K with 50% xylose as cryoprotectant [Bacillus thermoproteolyticus],6D5S_A Hexagonal thermolysin cryocooled to 100 K with 50% MPD as cryoprotectant [Bacillus thermoproteolyticus],6D5T_A Hexagonal thermolysin cryocooled to 100 K with 50% MPD as cryoprotectant [Bacillus thermoproteolyticus],6D5U_A Hexagonal thermolysin cryocooled to 100 K with 50% methanol as cryoprotectant [Bacillus thermoproteolyticus],6FJ2_A Structure of Thermolysin solved from SAD data collected at the peak of the Zn absorption edge on ID30B [Bacillus thermoproteolyticus],6GHX_A Alzheimer's Amyloid-Beta Peptide Fragment 31-35 in Complex with Cd-substituted Thermolysin [Geobacillus stearothermophilus],6IG7_A Crystal structure of thermolysin delivered in polyacrylamide using x-ray free electron laser [Bacillus thermoproteolyticus],6LZN_A Chain A, Thermolysin [Bacillus thermoproteolyticus],6LZO_A Chain A, Thermolysin [Bacillus thermoproteolyticus],6N4W_A Tetragonal thermolysin (with 50% xylose) cryocooled in a nitrogen gas stream to 100 K [Bacillus thermoproteolyticus],6N4Z_A Tetragonal thermolysin (with 50% xylose) plunge cooled in liquid nitrogen to 77 K [Bacillus thermoproteolyticus],6QAR_A Thermolysine under 2 kbar of argon [Bacillus thermoproteolyticus],6QF2_A Chain A, Thermolysin [Bacillus thermoproteolyticus],6QF3_A Chain A, Thermolysin [Bacillus thermoproteolyticus],6SB9_E Thermolysin in complex with J28 [Bacillus thermoproteolyticus],6SBK_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J13 [Bacillus thermoproteolyticus],6SC0_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J22 [Bacillus thermoproteolyticus],6SC1_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J96 [Bacillus thermoproteolyticus],6SC3_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J62 [Bacillus thermoproteolyticus],6SCK_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J77 [Bacillus thermoproteolyticus],6SCU_E THERMOLYSIN IN COMPLEX WITH FRAGMENT J88 [Bacillus thermoproteolyticus],6SEL_A Multicrystal structure of Thermolysin at room temperature using a multilayer monochromator. [Bacillus thermoproteolyticus],6Y4I_E Chain E, Thermolysin [Geobacillus stearothermophilus],6YI6_E Chain E, Thermolysin [Geobacillus stearothermophilus],6YMR_E Chain E, Thermolysin [Geobacillus stearothermophilus],6YMS_E Chain E, Thermolysin [Geobacillus stearothermophilus],6ZHJ_A Chain A, Thermolysin [Bacillus thermoproteolyticus],7AKN_A Thermolysin from Bacillus thermoproteolyticus [Bacillus thermoproteolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P43263 | 4.62e-188 | 39 | 525 | 41 | 526 | Bacillolysin OS=Brevibacillus brevis OX=1393 GN=npr PE=1 SV=1 |
| P29148 | 5.19e-182 | 14 | 524 | 13 | 590 | Bacillolysin OS=Paenibacillus polymyxa OX=1406 GN=npr PE=1 SV=1 |
| P05806 | 6.81e-146 | 39 | 525 | 45 | 565 | Bacillolysin OS=Bacillus cereus OX=1396 GN=npr PE=1 SV=2 |
| D5DEH5 | 6.29e-139 | 35 | 525 | 43 | 561 | Bacillolysin OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=nprM PE=3 SV=1 |
| P0CH29 | 6.97e-138 | 35 | 525 | 43 | 561 | Bacillolysin OS=Priestia megaterium (strain ATCC 14581 / DSM 32 / CCUG 1817 / JCM 2506 / NBRC 15308 / NCIMB 9376 / NCTC 10342 / NRRL B-14308 / VKM B-512 / Ford 19) OX=1348623 GN=nprM PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
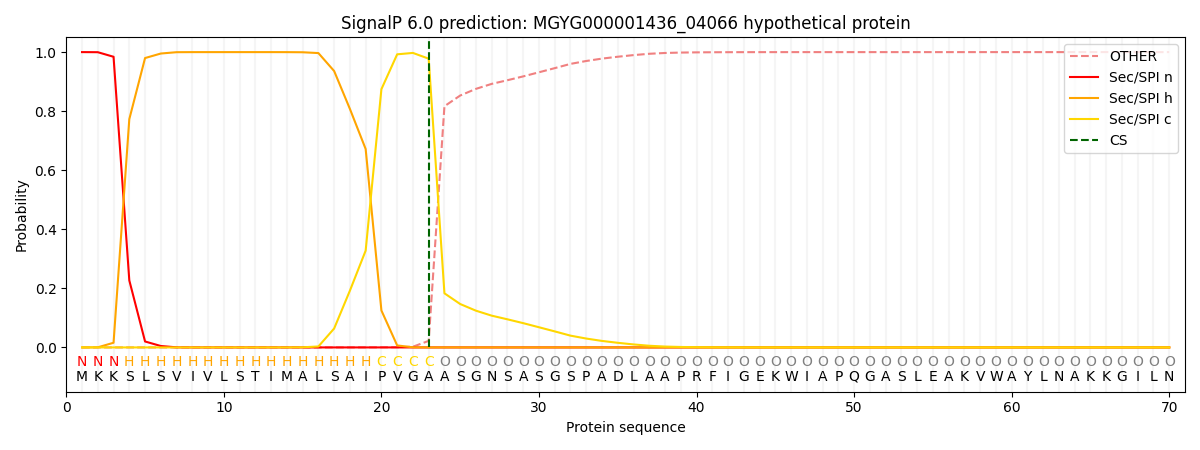
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999034 | 0.000158 | 0.000184 | 0.000161 | 0.000143 |
