You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001437_01658
You are here: Home > Sequence: MGYG000001437_01658
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus schleiferi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus schleiferi | |||||||||||
| CAZyme ID | MGYG000001437_01658 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 318392; End: 319654 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 92 | 368 | 1.6e-62 | 0.9619771863117871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.04e-25 | 107 | 368 | 26 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.38e-20 | 108 | 369 | 76 | 363 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG3934 | COG3934 | 0.001 | 104 | 383 | 25 | 293 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGS46457.1 | 0.0 | 1 | 420 | 1 | 420 |
| QPA24418.1 | 0.0 | 1 | 420 | 1 | 420 |
| QPA34648.1 | 0.0 | 1 | 420 | 1 | 420 |
| CAD7358579.1 | 5.15e-310 | 1 | 405 | 1 | 405 |
| AKS72584.1 | 5.16e-309 | 1 | 420 | 1 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3O6A_A | 3.15e-15 | 37 | 308 | 2 | 268 | F144Y/F258YDouble Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
| 4M80_A | 5.66e-15 | 37 | 308 | 2 | 268 | Thestructure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution [Candida albicans SC5314],4M81_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution [Candida albicans SC5314],4M82_A The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with p-nitrophenyl-gentiobioside (product) at 1.6A resolution [Candida albicans SC5314] |
| 2PB1_A | 5.69e-15 | 37 | 308 | 3 | 269 | Exo-B-(1,3)-Glucanasefrom Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
| 2PC8_A | 5.69e-15 | 37 | 308 | 3 | 269 | ChainA, Hypothetical protein XOG1 [Candida albicans] |
| 2PF0_A | 7.61e-15 | 37 | 308 | 3 | 269 | ChainA, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 3.69e-30 | 30 | 373 | 1 | 342 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| A1CRV0 | 4.10e-20 | 8 | 348 | 3 | 323 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=exgA PE=3 SV=2 |
| Q8NKF9 | 3.92e-18 | 33 | 300 | 21 | 287 | Glucan 1,3-beta-glucosidase OS=Candida oleophila OX=45573 GN=EXG1 PE=3 SV=1 |
| A2RAR6 | 1.21e-17 | 11 | 294 | 6 | 270 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
| Q96V64 | 2.33e-17 | 20 | 294 | 8 | 270 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
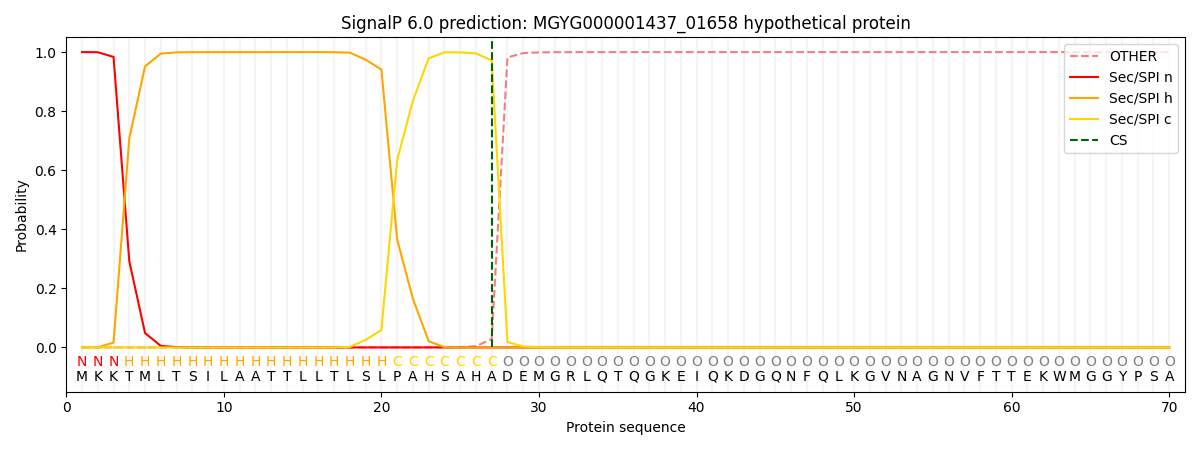
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000217 | 0.999036 | 0.000202 | 0.000195 | 0.000172 | 0.000151 |
