You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001438_01421
You are here: Home > Sequence: MGYG000001438_01421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
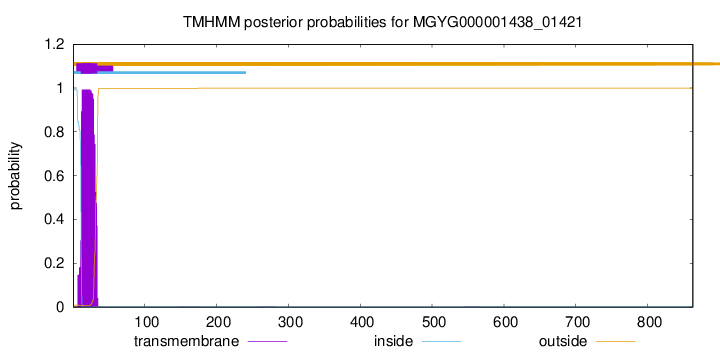
TMHMM annotations
Basic Information help
| Species | Paenisporosarcina sp000411295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_A; Planococcaceae; Paenisporosarcina; Paenisporosarcina sp000411295 | |||||||||||
| CAZyme ID | MGYG000001438_01421 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | 3',5'-cyclic adenosine monophosphate phosphodiesterase CpdA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1403382; End: 1405973 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM6 | 41 | 177 | 4.5e-26 | 0.8768115942028986 |
| CBM32 | 736 | 859 | 1.2e-24 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04084 | CBM6_xylanase-like | 3.12e-33 | 39 | 175 | 1 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| pfam03422 | CBM_6 | 1.71e-27 | 42 | 177 | 1 | 125 | Carbohydrate binding module (family 6). |
| smart00606 | CBD_IV | 1.48e-25 | 34 | 175 | 1 | 129 | Cellulose Binding Domain Type IV. |
| cd07402 | MPP_GpdQ | 1.50e-17 | 367 | 564 | 4 | 210 | Enterobacter aerogenes GpdQ and related proteins, metallophosphatase domain. GpdQ (glycerophosphodiesterase Q, also known as Rv0805 in Mycobacterium tuberculosis) is a binuclear metallophosphoesterase from Enterobacter aerogenes that catalyzes the hydrolysis of mono-, di-, and triester substrates, including some organophosphate pesticides and products of the degradation of nerve agents. The GpdQ homolog, Rv0805, has 2',3'-cyclic nucleotide phosphodiesterase activity. GpdQ and Rv0805 belong to the metallophosphatase (MPP) superfamily. MPPs are functionally diverse, but all share a conserved domain with an active site consisting of two metal ions (usually manganese, iron, or zinc) coordinated with octahedral geometry by a cage of histidine, aspartate, and asparagine residues. The MPP superfamily includes: Mre11/SbcD-like exonucleases, Dbr1-like RNA lariat debranching enzymes, YfcE-like phosphodiesterases, purple acid phosphatases (PAPs), YbbF-like UDP-2,3-diacylglucosamine hydrolases, and acid sphingomyelinases (ASMases). The conserved domain is a double beta-sheet sandwich with a di-metal active site made up of residues located at the C-terminal side of the sheets. This domain is thought to allow for productive metal coordination. |
| pfam00754 | F5_F8_type_C | 1.45e-16 | 732 | 858 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ98993.1 | 2.23e-42 | 724 | 863 | 296 | 434 |
| ABX42085.1 | 1.20e-32 | 722 | 861 | 311 | 454 |
| QNU66653.1 | 7.95e-30 | 718 | 861 | 813 | 953 |
| AIQ74597.1 | 1.02e-29 | 622 | 862 | 427 | 652 |
| SET83267.1 | 2.42e-29 | 402 | 602 | 390 | 615 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V4V_A | 5.37e-24 | 36 | 177 | 2 | 129 | CrystalStructure of a Family 6 Carbohydrate-Binding Module from Clostridium cellulolyticum in complex with xylose [Ruminiclostridium cellulolyticum] |
| 1K85_A | 5.00e-18 | 627 | 717 | 3 | 88 | ChainA, CHITINASE A1 [Niallia circulans] |
| 1UY1_A | 6.14e-17 | 36 | 157 | 19 | 129 | Bindingsub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY2_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY3_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY4_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium] |
| 1GMM_A | 6.36e-15 | 36 | 157 | 2 | 114 | ChainA, CBM6 [Acetivibrio thermocellus],1UXX_X Chain X, Xylanase U [Acetivibrio thermocellus] |
| 4FD0_A | 1.62e-12 | 179 | 256 | 26 | 103 | Crystalstructure of a putative cell surface protein (BACCAC_03700) from Bacteroides caccae ATCC 43185 at 2.07 A resolution [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 9.03e-16 | 624 | 719 | 556 | 646 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P33558 | 2.18e-14 | 26 | 157 | 232 | 358 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| Q8GJ44 | 3.03e-14 | 26 | 157 | 231 | 357 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P50899 | 1.29e-13 | 624 | 755 | 700 | 828 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
| P10478 | 5.80e-13 | 29 | 175 | 289 | 419 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
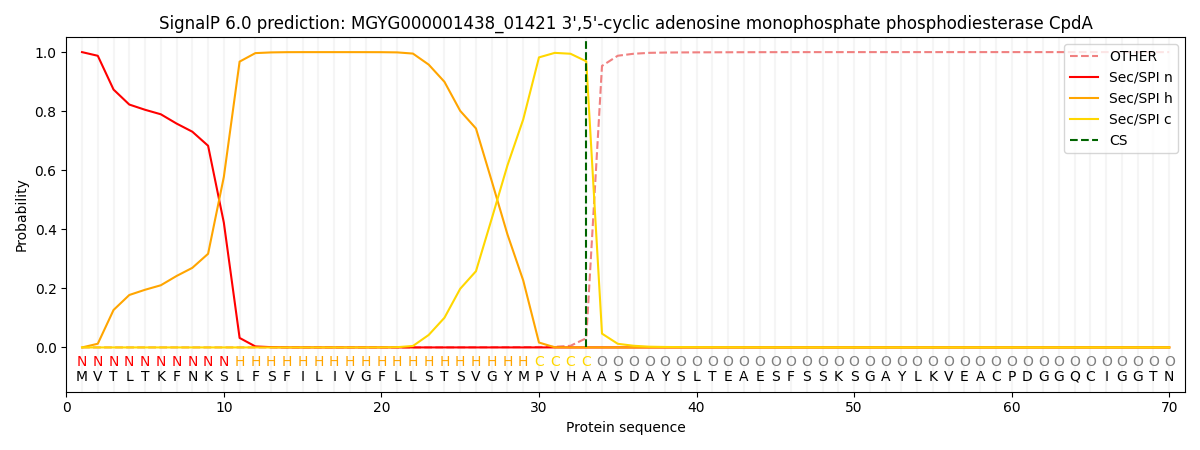
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000486 | 0.998793 | 0.000182 | 0.000190 | 0.000156 | 0.000144 |