You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001439_00277
You are here: Home > Sequence: MGYG000001439_00277
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
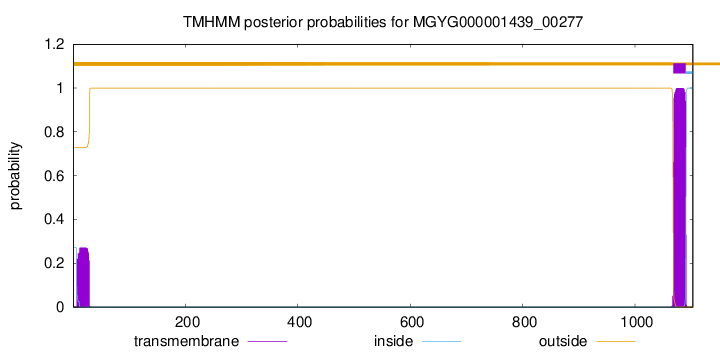
TMHMM annotations
Basic Information help
| Species | Faecalimonas umbilicata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas umbilicata | |||||||||||
| CAZyme ID | MGYG000001439_00277 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 272884; End: 276195 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 424 | 982 | 9.8e-161 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033839 | PspC_subgroup_2 | 2.36e-17 | 975 | 1057 | 417 | 497 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| NF033839 | PspC_subgroup_2 | 1.30e-16 | 988 | 1058 | 391 | 470 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| NF033839 | PspC_subgroup_2 | 2.27e-16 | 975 | 1060 | 340 | 430 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| NF033839 | PspC_subgroup_2 | 2.63e-16 | 988 | 1058 | 369 | 448 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| NF033839 | PspC_subgroup_2 | 5.37e-16 | 988 | 1058 | 336 | 415 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDS97815.1 | 9.27e-262 | 71 | 988 | 245 | 1190 |
| AIY03209.1 | 9.33e-262 | 68 | 989 | 223 | 1169 |
| QLL50162.1 | 2.08e-226 | 71 | 982 | 47 | 977 |
| QLL52630.1 | 2.08e-226 | 71 | 982 | 47 | 977 |
| ALA27928.1 | 2.08e-226 | 71 | 982 | 47 | 977 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 8.98e-63 | 425 | 985 | 10 | 579 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 1.78e-55 | 419 | 982 | 13 | 596 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 2.06e-53 | 425 | 987 | 15 | 613 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 4.09e-40 | 426 | 770 | 247 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 7.30e-40 | 426 | 770 | 247 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
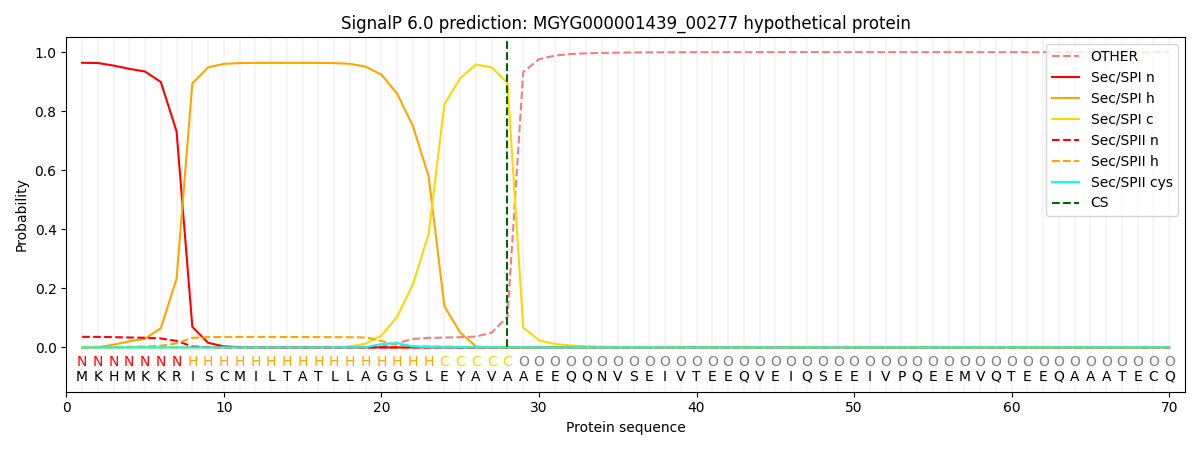
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001524 | 0.959785 | 0.037660 | 0.000497 | 0.000277 | 0.000221 |