You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001440_01996
You are here: Home > Sequence: MGYG000001440_01996
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acidaminococcus intestini | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Acidaminococcus; Acidaminococcus intestini | |||||||||||
| CAZyme ID | MGYG000001440_01996 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2133036; End: 2134673 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 3 | 478 | 2.3e-100 | 0.8925925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 9.64e-45 | 21 | 415 | 25 | 427 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 3.73e-36 | 31 | 372 | 33 | 375 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 2.65e-18 | 59 | 217 | 1 | 157 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEQ23377.1 | 0.0 | 1 | 545 | 1 | 545 |
| ADB46704.1 | 3.50e-264 | 1 | 545 | 1 | 546 |
| QTV78033.1 | 3.32e-134 | 1 | 525 | 1 | 512 |
| AXL22207.1 | 6.98e-107 | 12 | 540 | 39 | 517 |
| AVO27725.1 | 2.05e-103 | 7 | 543 | 29 | 508 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 2.06e-32 | 6 | 325 | 33 | 358 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A4SQX1 | 1.66e-33 | 25 | 366 | 24 | 367 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas salmonicida (strain A449) OX=382245 GN=arnT PE=3 SV=1 |
| B4EUL1 | 5.68e-33 | 1 | 333 | 5 | 335 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT1 PE=3 SV=1 |
| A0KGY4 | 7.63e-33 | 25 | 359 | 24 | 365 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 2.70e-32 | 9 | 372 | 8 | 374 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
| B4TBG8 | 1.10e-29 | 4 | 358 | 6 | 358 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella heidelberg (strain SL476) OX=454169 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.996029 | 0.003755 | 0.000194 | 0.000007 | 0.000006 | 0.000019 |
TMHMM Annotations download full data without filtering help
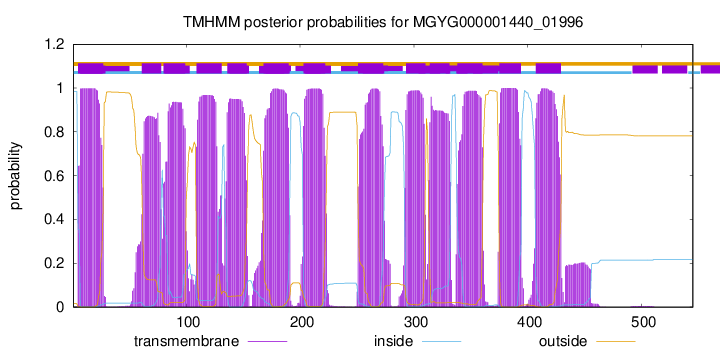
| start | end |
|---|---|
| 5 | 27 |
| 61 | 78 |
| 80 | 99 |
| 109 | 131 |
| 136 | 153 |
| 168 | 190 |
| 203 | 222 |
| 251 | 273 |
| 293 | 310 |
| 314 | 331 |
| 338 | 360 |
| 375 | 394 |
| 407 | 429 |
