You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001441_01600
You are here: Home > Sequence: MGYG000001441_01600
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp000411415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp000411415 | |||||||||||
| CAZyme ID | MGYG000001441_01600 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | putative dolichyl-phosphate-mannose--protein mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 612137; End: 613879 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 74 | 360 | 8.9e-66 | 0.9910313901345291 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1928 | PMT1 | 2.05e-17 | 80 | 580 | 36 | 694 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam16192 | PMT_4TMC | 3.87e-16 | 381 | 577 | 1 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| pfam13231 | PMT_2 | 2.42e-09 | 139 | 251 | 1 | 111 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG4346 | COG4346 | 2.44e-09 | 138 | 524 | 134 | 421 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
| COG1807 | ArnT | 2.93e-09 | 80 | 370 | 19 | 253 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGS11599.1 | 0.0 | 1 | 580 | 1 | 581 |
| QCT34948.1 | 0.0 | 1 | 580 | 1 | 581 |
| AKU65332.1 | 0.0 | 1 | 580 | 14 | 584 |
| QQC43974.1 | 0.0 | 1 | 580 | 14 | 584 |
| QWC16849.1 | 7.82e-128 | 67 | 580 | 116 | 622 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L8F4Z2 | 6.09e-66 | 65 | 579 | 35 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| P9WN04 | 4.03e-55 | 67 | 579 | 43 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 4.03e-55 | 67 | 579 | 43 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q8NRZ6 | 5.78e-47 | 50 | 579 | 21 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999697 | 0.000242 | 0.000048 | 0.000003 | 0.000001 | 0.000012 |
TMHMM Annotations download full data without filtering help
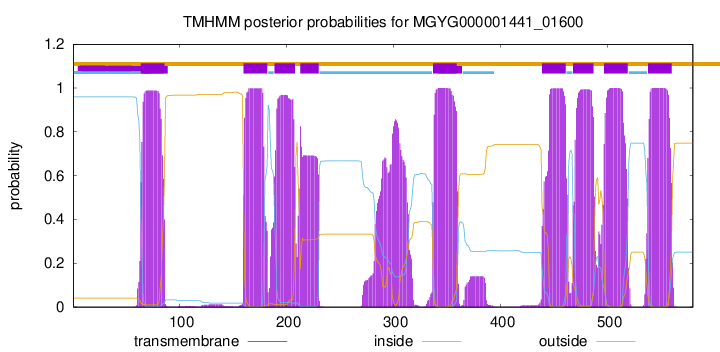
| start | end |
|---|---|
| 64 | 86 |
| 160 | 182 |
| 189 | 208 |
| 213 | 230 |
| 337 | 359 |
| 439 | 461 |
| 468 | 487 |
| 497 | 519 |
| 538 | 560 |
