You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_00025
You are here: Home > Sequence: MGYG000001443_00025
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_00025 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69320; End: 72193 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01804 | Penicil_amidase | 6.63e-106 | 123 | 944 | 1 | 626 | Penicillin amidase. Penicillin amidase or penicillin acylase EC:3.5.1.11 catalyzes the hydrolysis of benzylpenicillin to phenylacetic acid and 6-aminopenicillanic acid (6-APA) a key intermediate in the the synthesis of penicillins. Also in the family is cephalosporin acylase and aculeacin A acylase which are involved in the synthesis of related peptide antibiotics. |
| COG2366 | PvdQ | 4.04e-40 | 122 | 833 | 41 | 612 | Acyl-homoserine lactone (AHL) acylase PvdQ [Secondary metabolites biosynthesis, transport and catabolism]. |
| cd03748 | Ntn_PGA | 1.41e-34 | 425 | 821 | 1 | 363 | Penicillin G acylase (PGA) is the key enzyme in the industrial production of beta-lactam antibiotics. PGA hydrolyzes the side chain of penicillin G and related beta-lactam antibiotics releasing 6-amino penicillanic acid (6-APA), a building block in the production of semisynthetic penicillins. PGA is widely distributed among microorganisms, including bacteria, yeast and filamentous fungi but it's in vivo role remains unclear. |
| cd03747 | Ntn_PGA_like | 2.32e-29 | 425 | 723 | 1 | 278 | Penicillin G acylase (PGA) belongs to a family of beta-lactam acylases that includes cephalosporin acylase (CA) and aculeacin A acylase. PGA and CA are crucial for the production of backbone chemicals like 6-aminopenicillanic acid and 7-aminocephalosporanic acid (7-ACA), which can be used to synthesize semi-synthetic penicillins and cephalosporins, respectively. While both PGA and CA have a conserved Ntn (N-terminal nucleophile) hydrolase fold and the structural similarity at their active sites is very high, their sequence similarity is low. |
| cd01936 | Ntn_CA | 0.002 | 125 | 175 | 1 | 51 | Cephalosporin acylase (CA) belongs to a family of beta-lactam acylases that includes penicillin G acylase (PGA) and aculeacin A acylase. PGA and CA are crucial for the production of backbone chemicals like 6-aminopenicillanic acid and 7-aminocephalosporanic acid (7-ACA), which can be used to synthesize semi-synthetic penicillins and cephalosporins, respectively. While both PGA and CA have a conserved Ntn (N-terminal nucleophile) hydrolase fold and the structural similarity at their active sites is very high, their sequence similarity to other Ntn's is low. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKU19272.1 | 0.0 | 37 | 957 | 23 | 945 |
| ACU72119.1 | 0.0 | 35 | 957 | 48 | 972 |
| AHH94081.1 | 0.0 | 1 | 957 | 16 | 953 |
| CCH27971.1 | 0.0 | 37 | 957 | 53 | 942 |
| QUQ69355.1 | 0.0 | 28 | 957 | 26 | 943 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NVX_B | 7.76e-30 | 425 | 832 | 1 | 377 | Crystalstructure of penicillin G acylase from Bacillus sp. FJAT-27231 [Bacillus sp. FJAT-27231] |
| 7EA4_A | 3.91e-16 | 87 | 840 | 15 | 669 | ChainA, D-succinylase [Cupriavidus sp. P4-10-C],7EA4_B Chain B, D-succinylase [Cupriavidus sp. P4-10-C] |
| 7EBY_A | 6.76e-16 | 87 | 840 | 15 | 669 | ChainA, D-succinylase [Cupriavidus sp. P4-10-C],7EBY_B Chain B, D-succinylase [Cupriavidus sp. P4-10-C],7EBY_C Chain C, D-succinylase [Cupriavidus sp. P4-10-C],7EBY_D Chain D, D-succinylase [Cupriavidus sp. P4-10-C],7EBY_E Chain E, D-succinylase [Cupriavidus sp. P4-10-C],7EBY_F Chain F, D-succinylase [Cupriavidus sp. P4-10-C] |
| 3ML0_A | 1.79e-09 | 121 | 321 | 3 | 191 | ThermostablePenicillin G acylase from Alcaligenes faecalis in tetragonal form [Alcaligenes faecalis] |
| 3K3W_A | 1.82e-09 | 121 | 321 | 3 | 191 | ThermostablePenicillin G acylase from Alcaligenes faecalis in orthorhombic form [Alcaligenes faecalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31956 | 2.53e-39 | 115 | 870 | 28 | 676 | Penicillin G acylase OS=Rhizobium viscosum OX=1673 GN=pac PE=1 SV=1 |
| P07941 | 1.41e-16 | 122 | 813 | 32 | 643 | Penicillin G acylase OS=Kluyvera cryocrescens OX=580 GN=pac PE=1 SV=1 |
| P06875 | 3.77e-15 | 122 | 813 | 32 | 645 | Penicillin G acylase OS=Escherichia coli OX=562 GN=pac PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
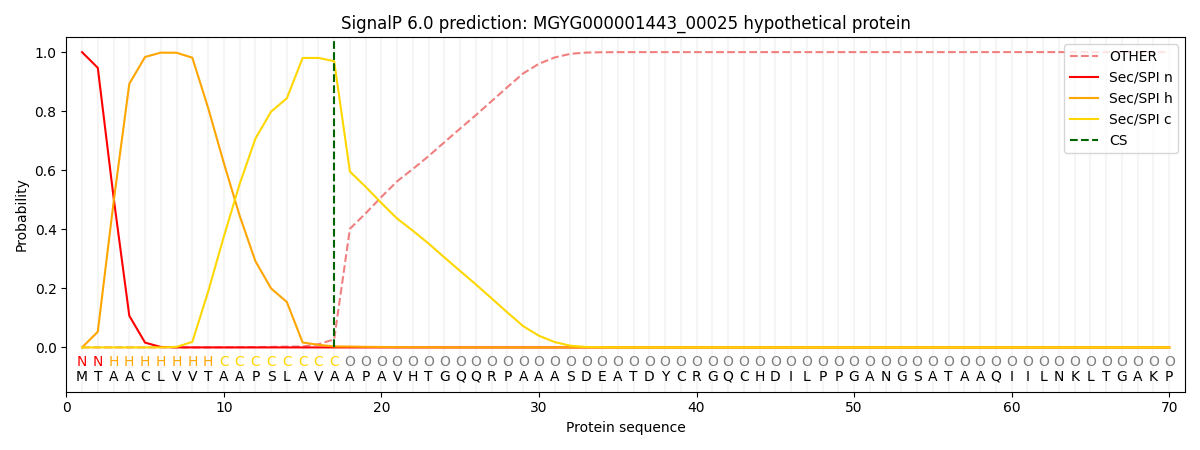
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001000 | 0.998092 | 0.000196 | 0.000259 | 0.000203 | 0.000198 |
