You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_01035
You are here: Home > Sequence: MGYG000001443_01035
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
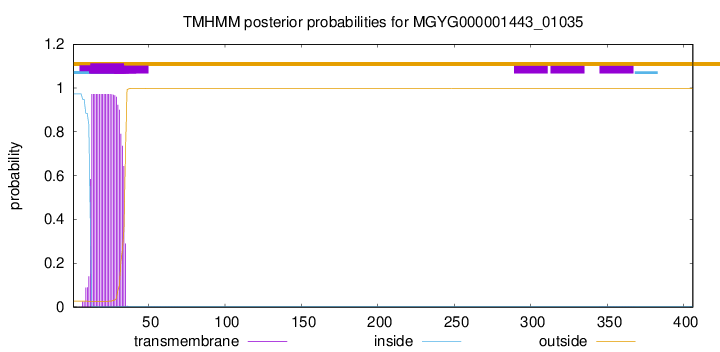
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_01035 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | Extracellular serine proteinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1335071; End: 1336291 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04077 | Peptidases_S8_PCSK9_ProteinaseK_like | 2.22e-116 | 139 | 385 | 1 | 255 | Peptidase S8 family domain in ProteinaseK-like proteins. The peptidase S8 or Subtilase clan of proteases have a Asp/His/Ser catalytic triad that is not homologous to trypsin. This CD contains several members of this clan including: PCSK9 (Proprotein convertase subtilisin/kexin type 9), Proteinase_K, Proteinase_T, and other subtilisin-like serine proteases. PCSK9 posttranslationally regulates hepatic low-density lipoprotein receptors (LDLRs) by binding to LDLRs on the cell surface, leading to their degradation. The binding site of PCSK9 has been localized to the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR. Characterized Proteinases K are secreted endopeptidases with a high degree of sequence conservation. Proteinases K are not substrate-specific and function in a wide variety of species in different pathways. It can hydrolyze keratin and other proteins with subtilisin-like specificity. The number of calcium-binding motifs found in these differ. Proteinase T is a novel proteinase from the fungus Tritirachium album Limber. The amino acid sequence of proteinase T as deduced from the nucleotide sequence is about 56% identical to that of proteinase K. |
| cd07484 | Peptidases_S8_Thermitase_like | 7.07e-69 | 156 | 385 | 25 | 258 | Peptidase S8 family domain in Thermitase-like proteins. Thermitase is a non-specific, trypsin-related serine protease with a very high specific activity. It contains a subtilisin like domain. The tertiary structure of thermitase is similar to that of subtilisin BPN'. It contains a Asp/His/Ser catalytic triad. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) clan include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53 the it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
| cd07477 | Peptidases_S8_Subtilisin_subset | 9.57e-61 | 165 | 380 | 6 | 227 | Peptidase S8 family domain in Subtilisin proteins. This group is composed of many different subtilisins: Pro-TK-subtilisin, subtilisin Carlsberg, serine protease Pb92 subtilisin, and BPN subtilisins just to name a few. Pro-TK-subtilisin is a serine protease from the hyperthermophilic archaeon Thermococcus kodakaraensis and consists of a signal peptide, a propeptide, and a mature domain. TK-subtilisin is matured from pro-TK-subtilisin upon autoprocessing and degradation of the propeptide. Unlike other subtilisins though, the folding of the unprocessed form of pro-TK-subtilisin is induced by Ca2+ binding which is almost completed prior to autoprocessing. Ca2+ is required for activity unlike the bacterial subtilisins. The propeptide is not required for folding of the mature domain unlike the bacterial subtilases because of the stability produced from Ca2+ binding. Subtilisin Carlsberg is extremely similar in structure to subtilisin BPN'/Novo thought it has a 30% difference in amino acid sequence. The substrate binding regions are also similar and 2 possible Ca2+ binding sites have been identified recently. Subtilisin Carlsberg possesses the highest commercial importance as a proteolytic additive for detergents. Serine protease Pb92, the serine protease from the alkalophilic Bacillus strain PB92, also contains two calcium ions and the overall folding of the polypeptide chain closely resembles that of the subtilisins. Members of the peptidases S8 and S35 clan include endopeptidases, exopeptidases and also a tripeptidyl-peptidase. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The S53 family contains a catalytic triad Glu/Asp/Ser. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07487 | Peptidases_S8_1 | 4.37e-52 | 158 | 384 | 1 | 264 | Peptidase S8 family domain, uncharacterized subfamily 1. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07473 | Peptidases_S8_Subtilisin_like | 8.07e-50 | 165 | 383 | 8 | 258 | Peptidase S8 family domain in Subtilisin-like proteins. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRK11067.1 | 2.72e-107 | 52 | 401 | 48 | 404 |
| AXO37891.1 | 1.98e-100 | 34 | 403 | 3 | 376 |
| AEB45483.1 | 5.69e-99 | 20 | 402 | 20 | 404 |
| ADO75815.1 | 7.24e-96 | 29 | 400 | 17 | 397 |
| CTQ90342.1 | 1.01e-91 | 9 | 403 | 6 | 391 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2B6N_A | 4.77e-100 | 133 | 400 | 3 | 274 | The1.8 A crystal structure of a Proteinase K like enzyme from a psychrotroph Serratia species [Serratia sp. GF96] |
| 1S2N_A | 9.41e-99 | 139 | 400 | 6 | 269 | Crystalstructure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1S2N_B Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1SH7_A Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44],1SH7_B Crystal structure of a cold adapted subtilisin-like serine proteinase [Vibrio sp. PA-44] |
| 5WSL_A | 3.10e-98 | 138 | 400 | 7 | 274 | Structuralstudies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220],5WSL_B Structural studies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220],5WSL_C Structural studies of keratinase from Meiothermus taiwanensis WR-220 [Meiothermus taiwanensis WR-220] |
| 4DZT_A | 9.75e-94 | 135 | 400 | 4 | 272 | AqualysinI: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1 [Thermus aquaticus] |
| 3F7M_A | 3.15e-63 | 132 | 399 | 2 | 273 | Crystalstructure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae [Lecanicillium psalliotae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P80146 | 1.70e-106 | 39 | 400 | 28 | 406 | Extracellular serine proteinase OS=Thermus sp. (strain Rt41A) OX=32063 PE=1 SV=3 |
| P16588 | 3.08e-100 | 51 | 400 | 53 | 412 | Alkaline serine exoprotease A OS=Vibrio alginolyticus OX=663 GN=proA PE=3 SV=1 |
| P08594 | 4.79e-100 | 22 | 400 | 13 | 399 | Aqualysin-1 OS=Thermus aquaticus OX=271 GN=pstI PE=1 SV=2 |
| P29138 | 6.53e-70 | 74 | 399 | 51 | 383 | Cuticle-degrading protease OS=Metarhizium anisopliae OX=5530 GN=PR1 PE=1 SV=1 |
| C5PGK9 | 7.35e-68 | 93 | 401 | 80 | 393 | Subtilisin-like protease CPC735_050320 OS=Coccidioides posadasii (strain C735) OX=222929 GN=CPC735_050320 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
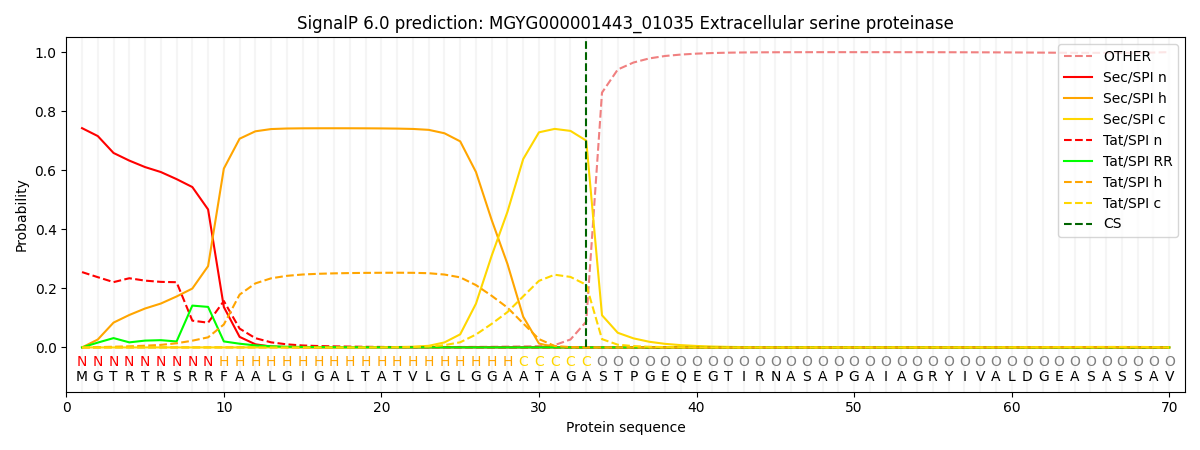
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001368 | 0.737547 | 0.002059 | 0.257831 | 0.000949 | 0.000228 |