You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_01151
Basic Information
help
| Species |
Streptomyces albus
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus
|
| CAZyme ID |
MGYG000001443_01151
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 721 |
|
72941.62 |
10.6107 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001443 |
7820353 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1451463;
End: 1453628
Strand: +
|
No EC number prediction in MGYG000001443_01151.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
33 |
486 |
6e-37 |
0.7888888888888889 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13231
|
PMT_2 |
1.84e-23 |
91 |
244 |
1 |
152 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1807
|
ArnT |
5.42e-16 |
88 |
265 |
60 |
234 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P37483
|
1.10e-49 |
32 |
402 |
4 |
444 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
|
O34575
|
1.59e-49 |
41 |
706 |
15 |
665 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999785
|
0.000241
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
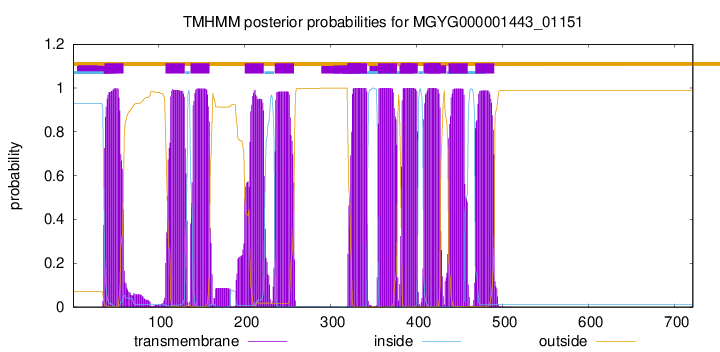
| start |
end |
| 37 |
59 |
| 108 |
130 |
| 137 |
159 |
| 200 |
222 |
| 235 |
257 |
| 320 |
342 |
| 355 |
377 |
| 382 |
401 |
| 408 |
427 |
| 437 |
459 |
| 468 |
490 |


