You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_01888
You are here: Home > Sequence: MGYG000001443_01888
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_01888 | |||||||||||
| CAZy Family | GT87 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2301947; End: 2304250 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT87 | 98 | 361 | 2.1e-46 | 0.9956709956709957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14378 | PAP2_3 | 2.78e-46 | 488 | 657 | 2 | 177 | PAP2 superfamily. |
| cd03386 | PAP2_Aur1_like | 2.60e-34 | 485 | 657 | 1 | 168 | PAP2_like proteins, Aur1_like subfamily. Yeast Aur1p or Ipc1p is necessary for the addition of inositol phosphate to ceramide, an essential step in yeast sphingolipid synthesis, and is the target of several antifungal compounds such as aureobasidin. |
| pfam09594 | GT87 | 5.01e-19 | 98 | 361 | 1 | 237 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
| PRK13375 | pimE | 7.32e-10 | 72 | 396 | 44 | 349 | mannosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID36178.1 | 0.0 | 1 | 767 | 1 | 767 |
| QHF93672.1 | 0.0 | 45 | 767 | 3 | 700 |
| QNF57540.1 | 0.0 | 45 | 767 | 3 | 700 |
| AXK34925.1 | 1.92e-300 | 43 | 676 | 1 | 607 |
| QNJ44611.1 | 2.39e-268 | 43 | 671 | 1 | 620 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WMZ8 | 8.42e-12 | 95 | 375 | 77 | 338 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2236 PE=3 SV=1 |
| P9WMZ9 | 8.42e-12 | 95 | 375 | 77 | 338 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv2181 PE=1 SV=1 |
| A0R036 | 1.24e-07 | 81 | 372 | 68 | 344 | Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=MSMEG_4247 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
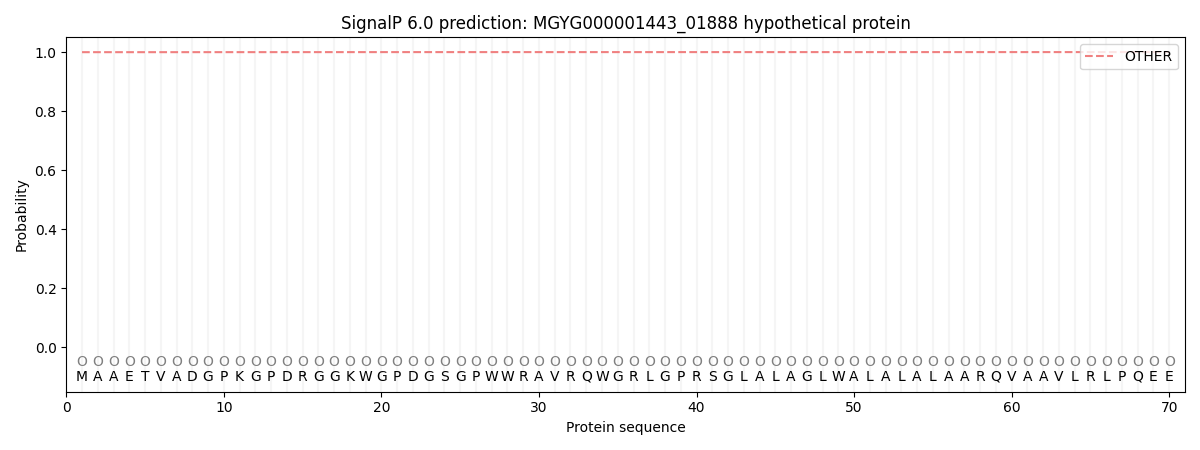
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999914 | 0.000102 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
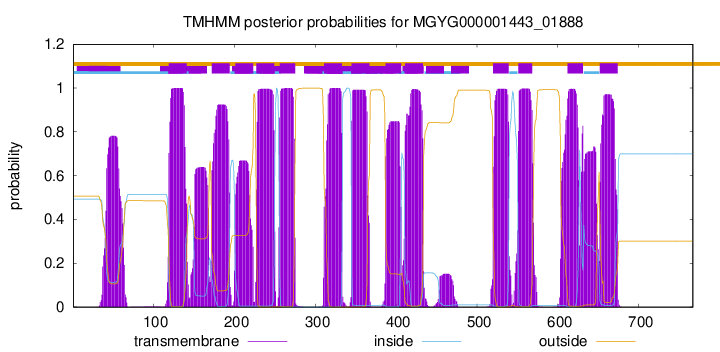
| start | end |
|---|---|
| 119 | 141 |
| 172 | 194 |
| 201 | 223 |
| 227 | 249 |
| 256 | 275 |
| 311 | 333 |
| 345 | 367 |
| 387 | 406 |
| 411 | 433 |
| 520 | 539 |
| 551 | 568 |
| 612 | 631 |
| 652 | 674 |
