You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_02196
You are here: Home > Sequence: MGYG000001443_02196
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
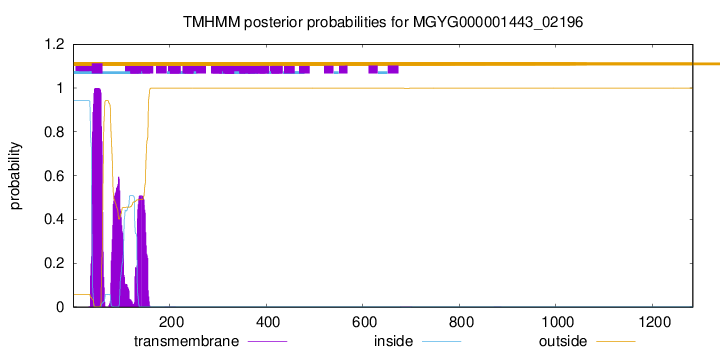
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_02196 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2654921; End: 2658775 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart01043 | BTAD | 6.51e-10 | 1125 | 1276 | 1 | 144 | Bacterial transcriptional activator domain. Found in the DNRI/REDD/AFSR family of regulators. This region of AFSR along with the C terminal region is capable of independently directing actinorhodin production. This family contains TPR repeats. |
| NF033845 | MSCRAMM_ClfB | 2.08e-07 | 418 | 617 | 540 | 743 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| NF033845 | MSCRAMM_ClfB | 2.32e-04 | 420 | 617 | 561 | 759 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| pfam04632 | FUSC | 3.60e-04 | 1076 | 1268 | 146 | 340 | Fusaric acid resistance protein family. This family includes a conserved region found in two proteins associated with fusaric acid resistance, FusC from Burkholderia cepacia and fdt-2 from Klebsiella oxytoca. These proteins are likely to be membrane transporter proteins. |
| COG3629 | DnrI | 0.001 | 1069 | 1268 | 41 | 226 | DNA-binding transcriptional activator of the SARP family [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXQ31232.1 | 1.08e-276 | 28 | 1283 | 23 | 974 |
| QWB23774.1 | 1.17e-267 | 30 | 1280 | 34 | 985 |
| QCN87429.1 | 1.85e-265 | 30 | 1280 | 27 | 971 |
| QNF63974.1 | 6.15e-264 | 30 | 1280 | 39 | 994 |
| QQN79321.1 | 2.57e-262 | 31 | 1283 | 13 | 1006 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
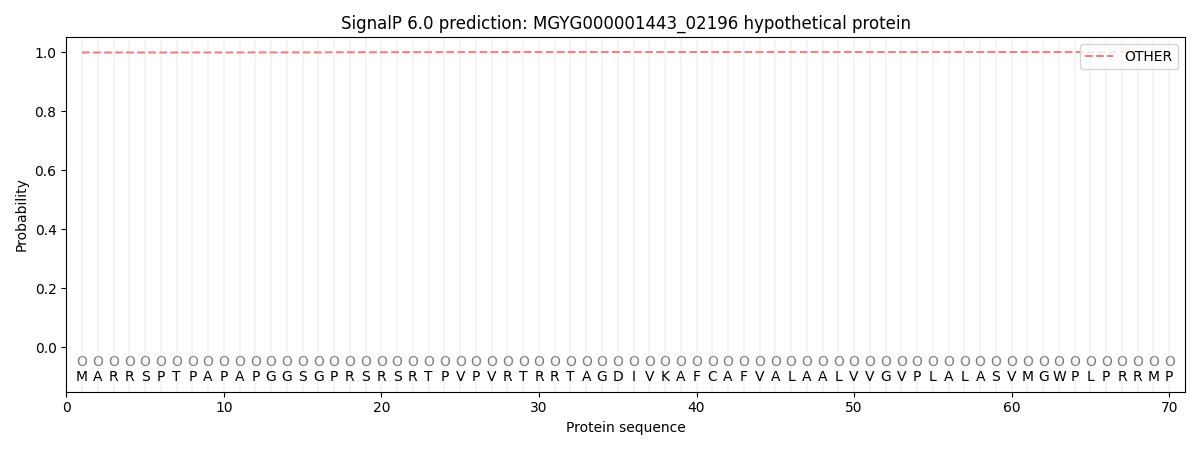
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999296 | 0.000726 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |