You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_02254
You are here: Home > Sequence: MGYG000001443_02254
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
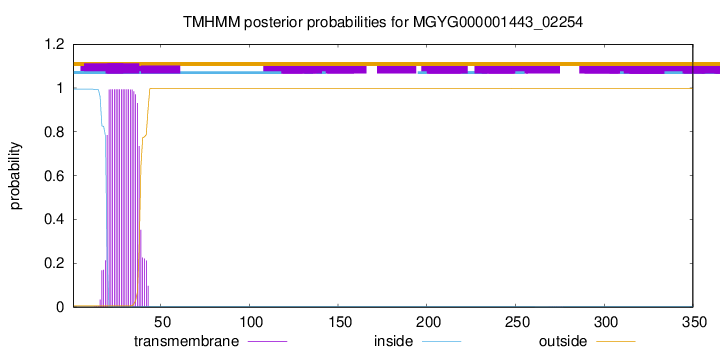
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_02254 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | putative bifunctional chitinase/lysozyme | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2725019; End: 2726071 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06543 | GH18_PF-ChiA-like | 6.86e-124 | 57 | 349 | 1 | 294 | PF-ChiA is an uncharacterized chitinase found in the hyperthermophilic archaeon Pyrococcus furiosus with a glycosyl hydrolase family 18 (GH18) catalytic domain as well as a cellulose-binding domain. Members of this domain family are found not only in archaea but also in eukaryotes and prokaryotes. PF-ChiA exhibits hydrolytic activity toward both colloidal and crystalline (beta/alpha) chitins at high temperature. |
| COG3469 | Chi1 | 8.03e-04 | 107 | 187 | 82 | 159 | Chitinase [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 0.001 | 115 | 174 | 65 | 124 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID36470.1 | 1.08e-229 | 1 | 350 | 1 | 350 |
| QHF93349.1 | 8.09e-155 | 55 | 350 | 64 | 359 |
| QNF57131.1 | 3.28e-154 | 55 | 350 | 64 | 359 |
| QPP08703.1 | 2.75e-134 | 55 | 350 | 56 | 351 |
| AKL64779.1 | 1.41e-133 | 52 | 348 | 196 | 491 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2DSK_A | 1.74e-35 | 59 | 348 | 12 | 289 | Crystalstructure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus],2DSK_B Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
| 3A4W_A | 9.17e-35 | 59 | 348 | 12 | 289 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4W_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus] |
| 3A4X_A | 2.49e-34 | 59 | 348 | 12 | 289 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4X_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3AFB_A Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus],3AFB_B Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13656 | 4.69e-45 | 59 | 348 | 589 | 891 | Probable bifunctional chitinase/lysozyme OS=Escherichia coli (strain K12) OX=83333 GN=chiA PE=1 SV=2 |
| O74199 | 5.27e-41 | 59 | 321 | 238 | 511 | Endochitinase 11 OS=Metarhizium anisopliae OX=5530 GN=chi11 PE=1 SV=1 |
| P14529 | 3.99e-40 | 80 | 350 | 64 | 323 | Chitinase OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=chiA2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
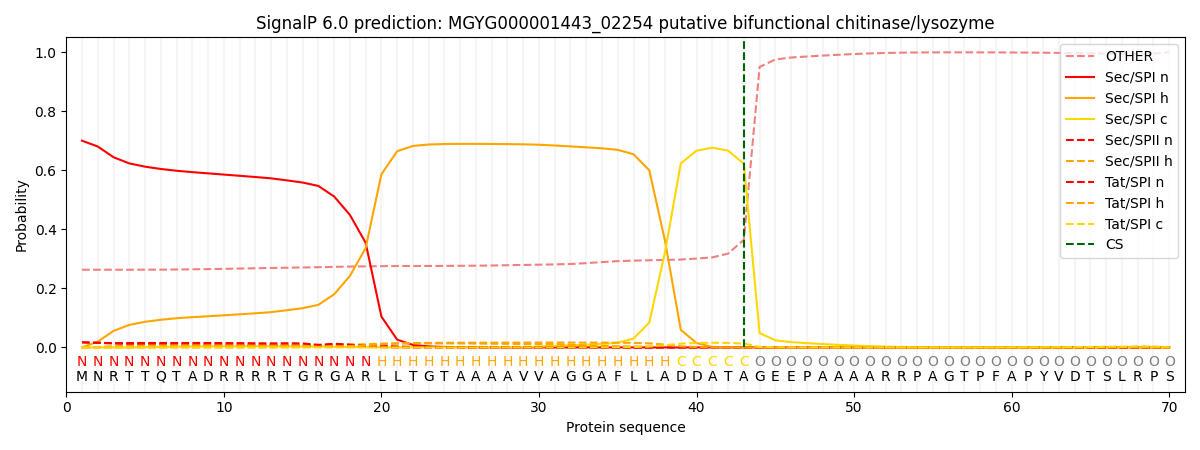
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.278563 | 0.678706 | 0.019372 | 0.019178 | 0.003523 | 0.000659 |