You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_02605
You are here: Home > Sequence: MGYG000001443_02605
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_02605 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Chitinase D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 166746; End: 167777 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 58 | 312 | 3.9e-24 | 0.6993243243243243 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02871 | GH18_chitinase_D-like | 1.07e-104 | 57 | 339 | 1 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| COG3469 | Chi1 | 9.14e-39 | 61 | 337 | 30 | 322 | Chitinase [Carbohydrate transport and metabolism]. |
| pfam00704 | Glyco_hydro_18 | 1.61e-22 | 61 | 325 | 7 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 3.81e-21 | 59 | 327 | 2 | 288 | Glyco_18 domain. |
| cd02877 | GH18_hevamine_XipI_class_III | 8.57e-18 | 57 | 330 | 1 | 273 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID40322.1 | 1.05e-231 | 26 | 343 | 1 | 318 |
| QHF98922.1 | 1.21e-196 | 11 | 343 | 4 | 337 |
| QNF59632.1 | 1.21e-196 | 11 | 343 | 4 | 337 |
| QPP10596.1 | 1.73e-175 | 51 | 343 | 34 | 327 |
| QKV97606.1 | 3.28e-168 | 51 | 343 | 14 | 307 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EBV_A | 2.32e-135 | 57 | 343 | 5 | 294 | Crystalstructure of putative Chitinase A from Streptomyces coelicolor. [Streptomyces coelicolor] |
| 5KZ6_A | 3.07e-50 | 59 | 343 | 12 | 332 | 1.25Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis],5KZ6_B 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis] |
| 4TX8_A | 1.75e-48 | 57 | 336 | 107 | 413 | CrystalStructure of a Family GH18 Chitinase from Chromobacterium violaceum [Chromobacterium violaceum ATCC 12472] |
| 3N11_A | 6.54e-48 | 59 | 334 | 9 | 320 | Crystalstricture of wild-type chitinase from Bacillus cereus NCTU2 [Bacillus cereus],3N12_A Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N15_A | 1.81e-47 | 59 | 334 | 9 | 320 | Crystalstricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 4.55e-47 | 61 | 343 | 194 | 514 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| Q05638 | 4.66e-44 | 45 | 335 | 252 | 591 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
| D4AVJ0 | 2.89e-39 | 74 | 325 | 16 | 313 | Probable class II chitinase ARB_00204 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00204 PE=1 SV=2 |
| A5FB63 | 1.33e-24 | 54 | 328 | 1139 | 1465 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| E9F7R6 | 6.89e-20 | 61 | 336 | 45 | 341 | Endochitinase 4 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
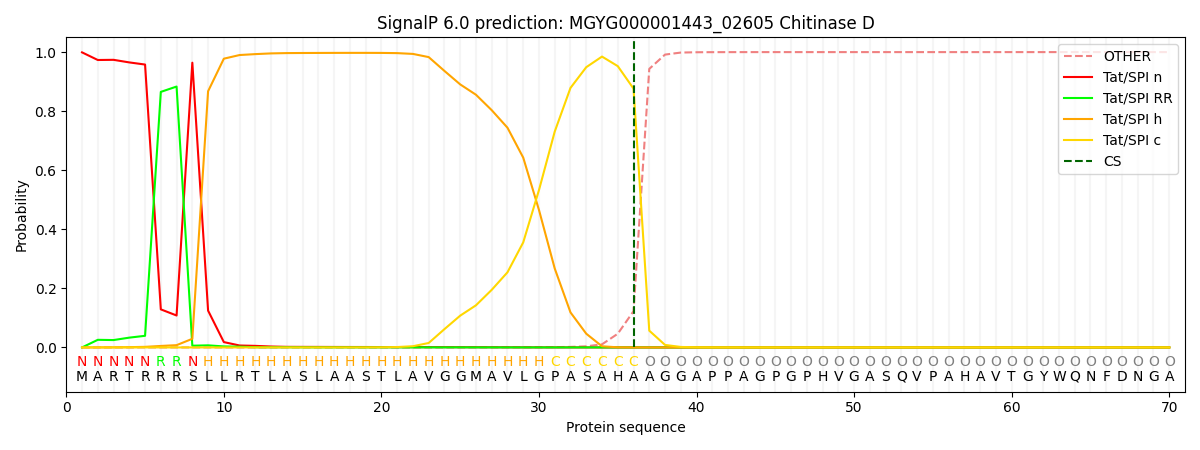
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000869 | 0.000000 | 0.999126 | 0.000002 | 0.000000 |

