You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_02627
You are here: Home > Sequence: MGYG000001443_02627
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
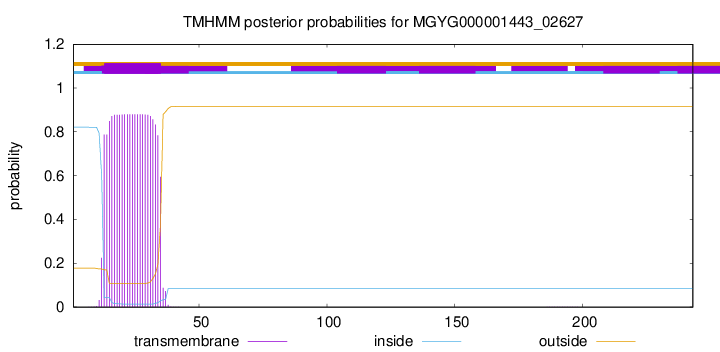
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_02627 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 188689; End: 189420 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01464 | SLT | 9.93e-05 | 175 | 234 | 22 | 85 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd13402 | LT_TF-like | 0.001 | 171 | 234 | 7 | 79 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13925 | RPF | 0.004 | 171 | 214 | 6 | 48 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID37977.1 | 5.65e-122 | 8 | 243 | 2 | 237 |
| QNF55444.1 | 1.63e-89 | 8 | 243 | 2 | 242 |
| QHF97170.1 | 1.63e-89 | 8 | 243 | 2 | 242 |
| QUI33610.1 | 1.01e-84 | 1 | 243 | 1 | 248 |
| AYA17004.1 | 1.11e-84 | 1 | 243 | 4 | 251 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.495235 | 0.409168 | 0.071540 | 0.015921 | 0.006319 | 0.001807 |