You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_03671
You are here: Home > Sequence: MGYG000001443_03671
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_03671 | |||||||||||
| CAZy Family | GT76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1426927; End: 1428204 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 91 | 276 | 1.4e-23 | 0.44963144963144963 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04188 | Mannosyl_trans2 | 3.99e-05 | 93 | 214 | 53 | 175 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJT63078.3 | 1.23e-191 | 80 | 425 | 1 | 346 |
| QSS91507.1 | 2.77e-186 | 59 | 425 | 1 | 377 |
| QRV56160.1 | 1.12e-185 | 59 | 425 | 1 | 377 |
| QLG32847.1 | 1.85e-184 | 59 | 425 | 1 | 377 |
| QES17089.1 | 5.82e-183 | 51 | 425 | 31 | 405 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7TPN3 | 4.09e-09 | 74 | 253 | 40 | 214 | GPI mannosyltransferase 2 OS=Mus musculus OX=10090 GN=Pigv PE=2 SV=2 |
| Q5KR61 | 6.88e-08 | 52 | 241 | 4 | 207 | GPI mannosyltransferase 2 OS=Rattus norvegicus OX=10116 GN=Pigv PE=2 SV=1 |
| Q9NUD9 | 3.74e-07 | 93 | 241 | 59 | 207 | GPI mannosyltransferase 2 OS=Homo sapiens OX=9606 GN=PIGV PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
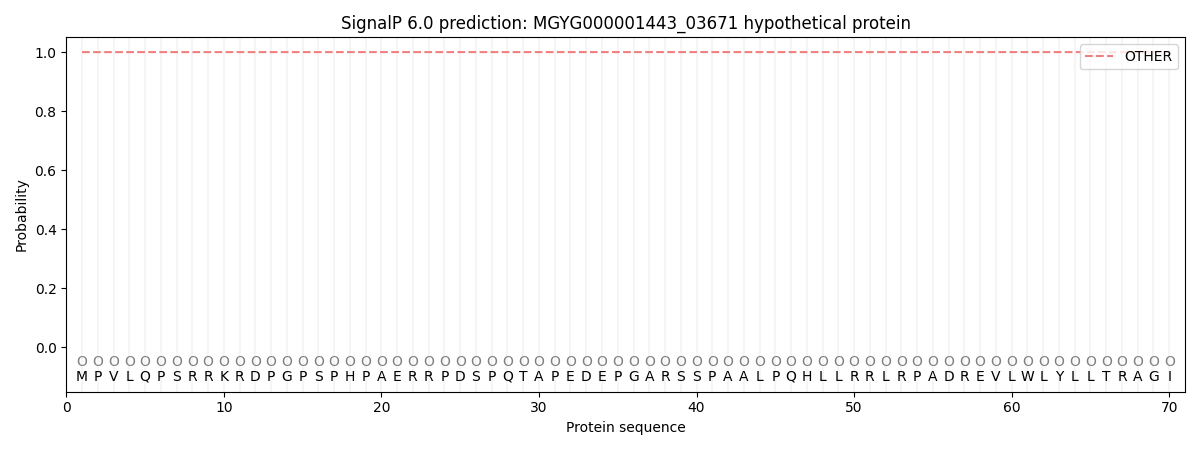
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999955 | 0.000068 | 0.000004 | 0.000000 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
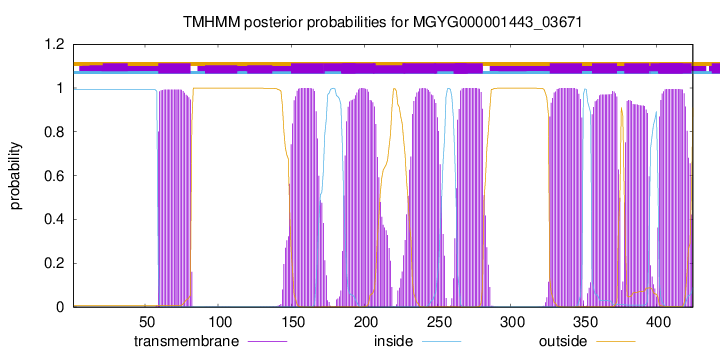
| start | end |
|---|---|
| 59 | 81 |
| 150 | 172 |
| 185 | 207 |
| 227 | 249 |
| 262 | 281 |
| 327 | 349 |
| 356 | 373 |
| 378 | 395 |
| 402 | 424 |
