You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_03870
You are here: Home > Sequence: MGYG000001443_03870
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_03870 | |||||||||||
| CAZy Family | GH99 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1650102; End: 1651478 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06283 | ThuA | 1.05e-111 | 54 | 265 | 1 | 213 | Trehalose utilisation. This family consists of several bacterial ThuA like proteins. ThuA appears to be involved in utilisation of trehalose. The thuA and thuB genes form part of the trehalose/sucrose transport operon thuEFGKAB, which is located on the pSymB megaplasmid. The thuA and thuB genes are induced in vitro by trehalose but not by sucrose and the extent of its induction depends on the concentration of trehalose available in the medium. |
| pfam06439 | DUF1080 | 3.08e-45 | 278 | 455 | 1 | 182 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| COG3828 | COG3828 | 8.54e-12 | 76 | 288 | 25 | 237 | Type 1 glutamine amidotransferase (GATase1)-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGJ52858.1 | 1.32e-228 | 14 | 457 | 11 | 459 |
| CQR65327.1 | 8.13e-228 | 54 | 455 | 49 | 450 |
| AJP00905.1 | 3.29e-227 | 54 | 455 | 49 | 450 |
| CAD5913137.1 | 1.96e-226 | 9 | 457 | 2 | 452 |
| CAD5994367.1 | 1.96e-226 | 9 | 457 | 2 | 452 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JQS_A | 3.34e-20 | 77 | 271 | 43 | 244 | Crystalstructure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 2.30 A resolution [Bacteroides uniformis ATCC 8492],4JQS_B Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 2.30 A resolution [Bacteroides uniformis ATCC 8492],4JQS_C Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 2.30 A resolution [Bacteroides uniformis ATCC 8492],4PXY_A Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution [Bacteroides uniformis ATCC 8492],4PXY_B Crystal structure of a Putative thua-like protein (BACUNI_01602) from Bacteroides uniformis ATCC 8492 at 1.50 A resolution [Bacteroides uniformis ATCC 8492] |
| 3IMM_A | 5.43e-08 | 323 | 457 | 46 | 201 | Crystalstructure of Putative glycosyl hydrolase (YP_001301887.1) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution [Parabacteroides distasonis ATCC 8503],3IMM_B Crystal structure of Putative glycosyl hydrolase (YP_001301887.1) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution [Parabacteroides distasonis ATCC 8503],3IMM_C Crystal structure of Putative glycosyl hydrolase (YP_001301887.1) from Parabacteroides distasonis ATCC 8503 at 2.00 A resolution [Parabacteroides distasonis ATCC 8503] |
| 4E5V_A | 1.67e-06 | 84 | 269 | 51 | 258 | Crystalstructure of a Putative thua-like protein (PARMER_02418) from Parabacteroides merdae ATCC 43184 at 1.75 A resolution [Parabacteroides merdae ATCC 43184],4E5V_B Crystal structure of a Putative thua-like protein (PARMER_02418) from Parabacteroides merdae ATCC 43184 at 1.75 A resolution [Parabacteroides merdae ATCC 43184] |
| 4HXC_A | 4.76e-06 | 278 | 458 | 35 | 263 | Crystalstructure of a putative glycosyl hydrolase (BACUNI_00951) from Bacteroides uniformis ATCC 8492 at 2.15 A resolution [Bacteroides uniformis ATCC 8492] |
| 3S5Q_A | 5.65e-06 | 277 | 457 | 18 | 214 | Crystalstructure of a putative glycosyl hydrolase (BDI_2473) from Parabacteroides distasonis ATCC 8503 at 1.85 A resolution [Parabacteroides distasonis ATCC 8503] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
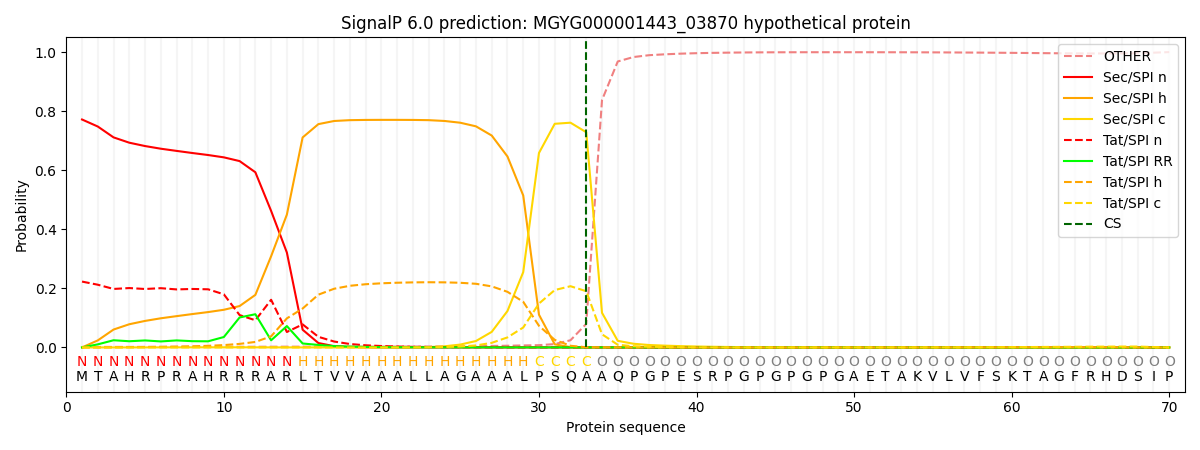
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003819 | 0.763950 | 0.003211 | 0.226852 | 0.001827 | 0.000295 |

