You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_04002
You are here: Home > Sequence: MGYG000001443_04002
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
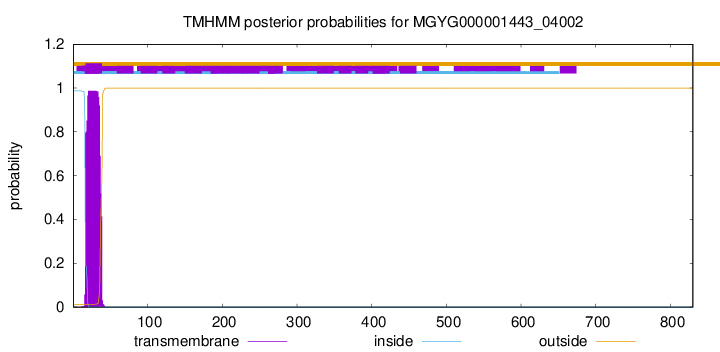
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_04002 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1815511; End: 1818003 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06283 | ThuA | 1.79e-77 | 59 | 279 | 1 | 213 | Trehalose utilisation. This family consists of several bacterial ThuA like proteins. ThuA appears to be involved in utilisation of trehalose. The thuA and thuB genes form part of the trehalose/sucrose transport operon thuEFGKAB, which is located on the pSymB megaplasmid. The thuA and thuB genes are induced in vitro by trehalose but not by sucrose and the extent of its induction depends on the concentration of trehalose available in the medium. |
| pfam07995 | GSDH | 2.88e-20 | 314 | 610 | 3 | 217 | Glucose / Sorbosone dehydrogenase. Members of this family are glucose/sorbosone dehydrogenases that possess a beta-propeller fold. |
| COG2133 | YliI | 8.26e-11 | 384 | 632 | 110 | 302 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| COG3828 | COG3828 | 1.31e-06 | 135 | 281 | 73 | 216 | Type 1 glutamine amidotransferase (GATase1)-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVT37843.1 | 9.99e-288 | 55 | 816 | 80 | 980 |
| AYY15444.1 | 3.27e-287 | 56 | 814 | 53 | 951 |
| AVT31684.1 | 5.63e-287 | 55 | 816 | 80 | 980 |
| AJE43219.1 | 1.18e-285 | 55 | 818 | 69 | 968 |
| QEV41720.1 | 1.18e-285 | 55 | 818 | 69 | 968 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2ISM_A | 3.44e-11 | 386 | 621 | 72 | 259 | Crystalstructure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 [Thermus thermophilus HB8],2ISM_B Crystal structure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 [Thermus thermophilus HB8] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
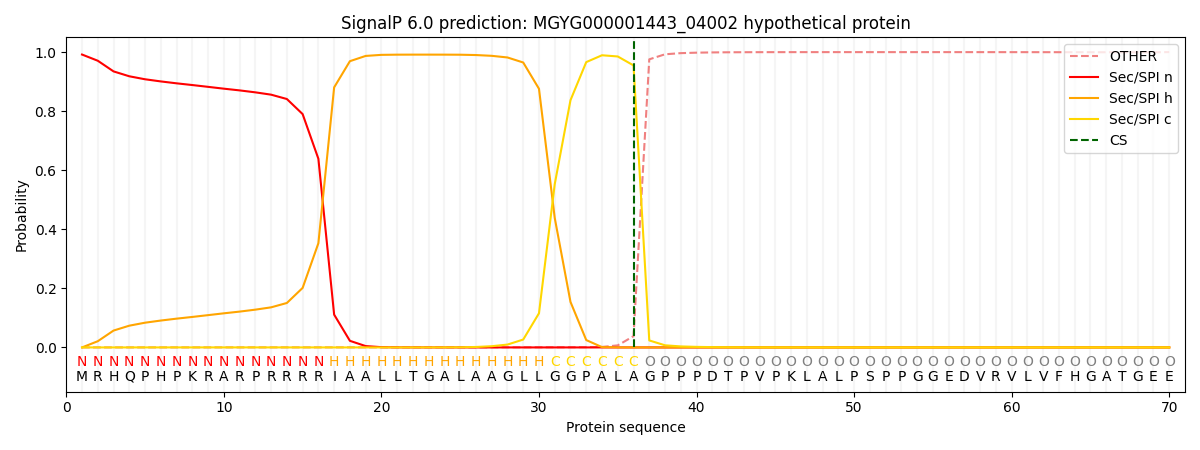
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000960 | 0.989329 | 0.000371 | 0.008808 | 0.000298 | 0.000198 |