You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_04020
You are here: Home > Sequence: MGYG000001443_04020
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_04020 | |||||||||||
| CAZy Family | GH114 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1839000; End: 1839872 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH114 | 80 | 252 | 6e-49 | 0.8578947368421053 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03537 | Glyco_hydro_114 | 4.45e-56 | 61 | 279 | 1 | 221 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG3868 | COG3868 | 5.07e-10 | 88 | 236 | 100 | 248 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID39059.1 | 4.07e-213 | 1 | 290 | 1 | 290 |
| QEU97893.1 | 1.08e-109 | 23 | 290 | 9 | 284 |
| QWQ45328.1 | 3.30e-109 | 21 | 290 | 24 | 286 |
| ARX83708.1 | 3.45e-109 | 16 | 290 | 7 | 277 |
| QEV70973.1 | 1.64e-108 | 17 | 285 | 12 | 287 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6OJ1_A | 5.06e-11 | 89 | 240 | 78 | 218 | CrystalStructure of Aspergillus fumigatus Ega3 [Aspergillus fumigatus Af293] |
| 6OJB_A | 5.76e-11 | 89 | 240 | 95 | 235 | CrystalStructure of Aspergillus fumigatus Ega3 complex with galactosamine [Aspergillus fumigatus Af293] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
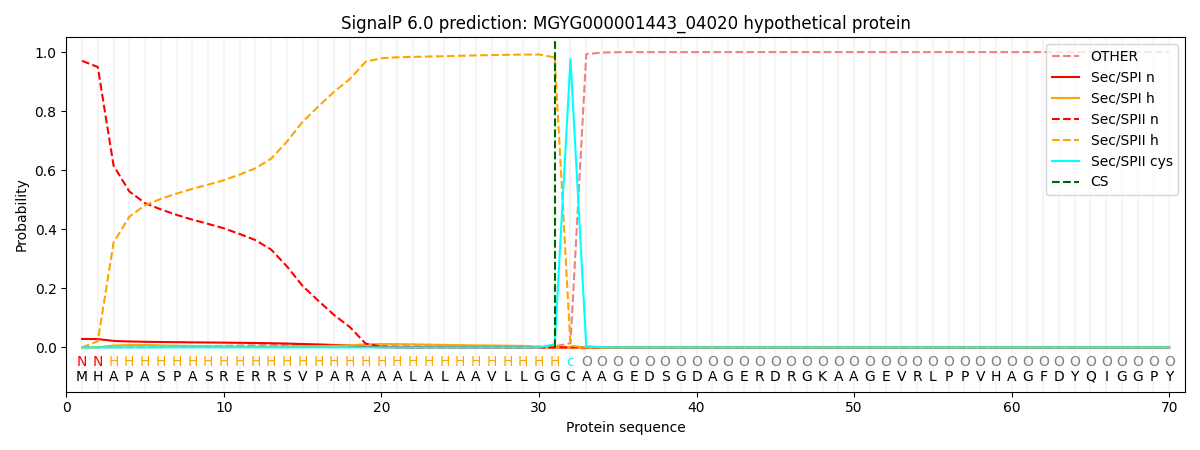
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000163 | 0.010390 | 0.989423 | 0.000029 | 0.000015 | 0.000003 |
