You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_04226
You are here: Home > Sequence: MGYG000001443_04226
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
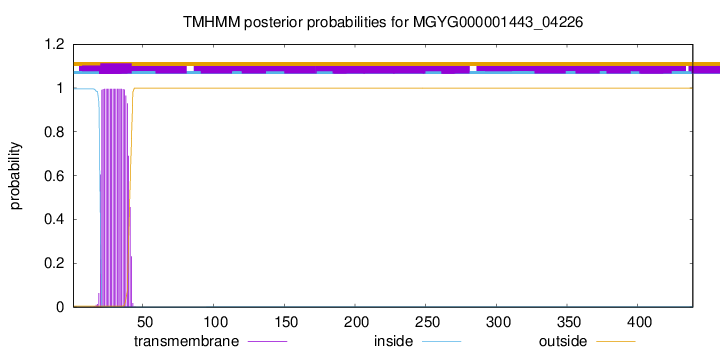
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_04226 | |||||||||||
| CAZy Family | GH0 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2063120; End: 2064439 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 5.72e-08 | 201 | 339 | 189 | 318 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 3.04e-04 | 201 | 272 | 160 | 232 | Glycosyl hydrolase family 26. |
| cd19100 | AKR_unchar | 0.002 | 177 | 272 | 141 | 232 | uncharacterized aldo-keto reductase (AKR) superfamily protein. This family includes a group of uncharacterized AKR superfamily proteins. Aldo-keto reductases (AKRs) are a superfamily of soluble NAD(P)(H) oxidoreductases whose chief purpose is to reduce aldehydes and ketones to primary and secondary alcohols. AKRs are present in all phyla and are of importance in both health and industrial applications. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID39224.1 | 1.29e-313 | 1 | 439 | 1 | 435 |
| QPP07141.1 | 4.03e-161 | 10 | 434 | 1 | 426 |
| AGS69695.1 | 6.19e-160 | 82 | 368 | 65 | 360 |
| AWT43373.1 | 1.43e-159 | 13 | 368 | 7 | 348 |
| AXE86006.1 | 1.88e-159 | 82 | 368 | 85 | 371 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
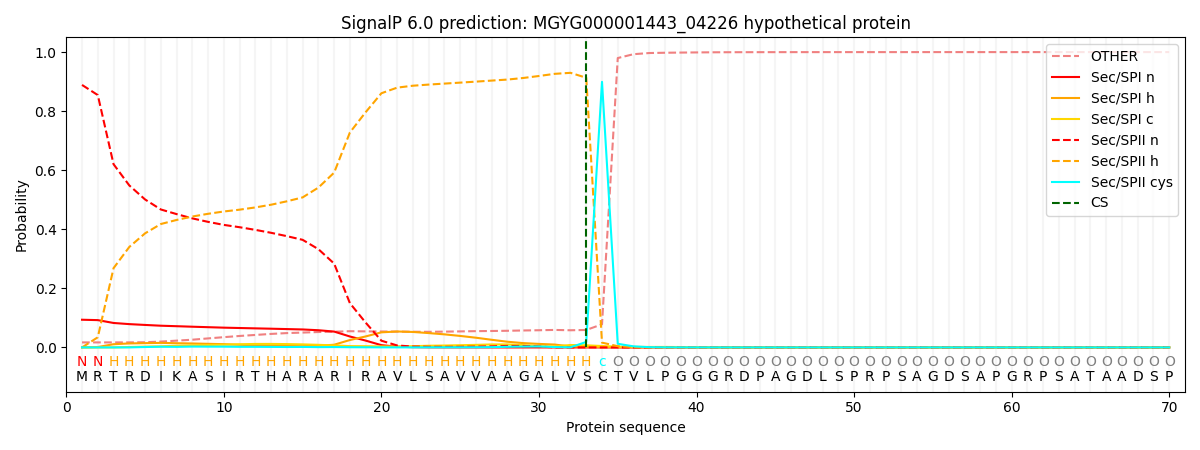
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.015973 | 0.034572 | 0.949221 | 0.000043 | 0.000190 | 0.000005 |