You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_05097
You are here: Home > Sequence: MGYG000001443_05097
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
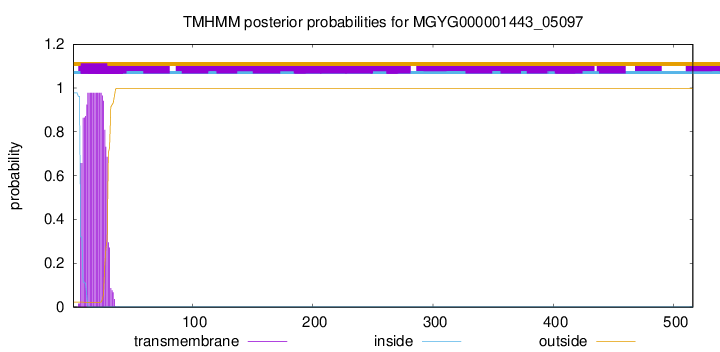
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_05097 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 225248; End: 226798 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01823 | SEST_like | 3.46e-30 | 81 | 353 | 3 | 253 | SEST_like. A family of secreted SGNH-hydrolases similar to Streptomyces scabies esterase (SEST), a causal agent of the potato scab disease, which hydrolyzes a specific ester bond in suberin, a plant lipid. The tertiary fold of this enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles two of the three components of typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxylic acid. |
| smart00458 | RICIN | 9.99e-11 | 400 | 515 | 6 | 116 | Ricin-type beta-trefoil. Carbohydrate-binding domain formed from presumed gene triplication. |
| pfam00652 | Ricin_B_lectin | 1.50e-10 | 393 | 475 | 46 | 126 | Ricin-type beta-trefoil lectin domain. |
| cd00161 | RICIN | 8.61e-10 | 355 | 475 | 12 | 122 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| pfam00652 | Ricin_B_lectin | 1.04e-09 | 394 | 514 | 4 | 126 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID37301.1 | 0.0 | 1 | 516 | 1 | 516 |
| QHF98009.1 | 4.45e-281 | 1 | 514 | 1 | 508 |
| QNF56208.1 | 4.45e-281 | 1 | 514 | 1 | 508 |
| AZS72773.1 | 4.45e-249 | 49 | 514 | 39 | 502 |
| QIK07901.1 | 2.46e-246 | 27 | 514 | 25 | 512 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HYQ_A | 6.26e-10 | 82 | 348 | 6 | 223 | Crystalstructure of phospholipase A1 from Streptomyces albidoflavus NA297 [Streptomyces albidoflavus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
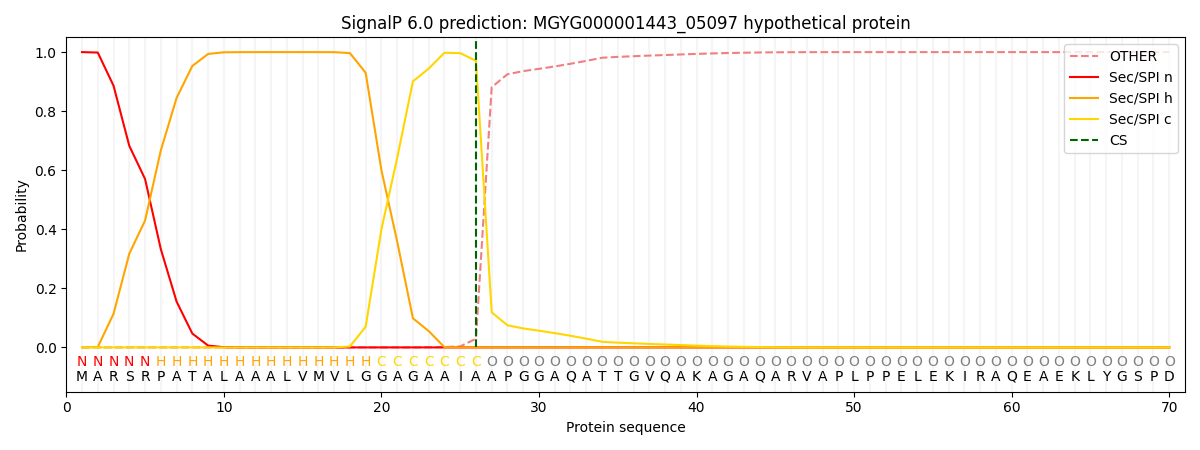
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000280 | 0.999042 | 0.000153 | 0.000190 | 0.000154 | 0.000142 |