You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_05431
You are here: Home > Sequence: MGYG000001443_05431
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
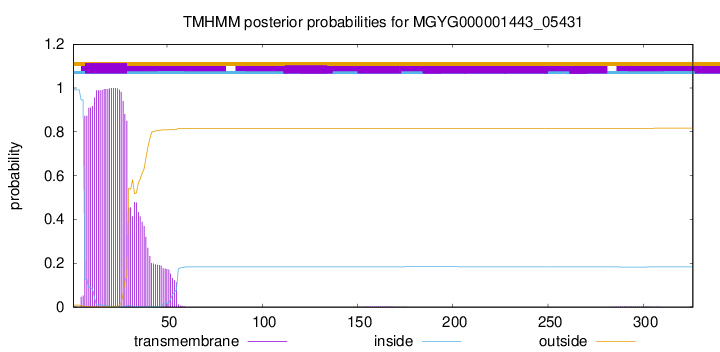
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_05431 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 618856; End: 619836 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 66 | 190 | 6.3e-30 | 0.8444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13399 | Slt35-like | 6.47e-37 | 69 | 188 | 3 | 108 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam00877 | NLPC_P60 | 3.93e-31 | 214 | 322 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 9.90e-30 | 181 | 323 | 54 | 193 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| cd00254 | LT-like | 6.35e-27 | 71 | 184 | 1 | 108 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| PRK13914 | PRK13914 | 9.34e-23 | 197 | 312 | 361 | 466 | invasion associated endopeptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNF56063.1 | 1.10e-190 | 3 | 325 | 5 | 328 |
| QHF97822.1 | 4.49e-190 | 3 | 325 | 5 | 328 |
| QEV22383.1 | 4.14e-176 | 4 | 322 | 2 | 320 |
| AZM57740.1 | 3.39e-175 | 4 | 322 | 2 | 320 |
| QES39142.1 | 4.58e-173 | 4 | 322 | 2 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CFL_A | 1.74e-22 | 206 | 306 | 18 | 115 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 3H41_A | 3.47e-18 | 198 | 317 | 185 | 299 | CRYSTALSTRUCTURE OF A NLPC/P60 FAMILY PROTEIN (BCE_2878) FROM BACILLUS CEREUS ATCC 10987 AT 1.79 A RESOLUTION [Bacillus cereus ATCC 10987] |
| 3PBI_A | 3.47e-17 | 195 | 312 | 78 | 199 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
| 3I86_A | 9.29e-17 | 211 | 315 | 21 | 129 | Crystalstructure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis],3I86_B Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis] |
| 3NE0_A | 3.23e-16 | 206 | 314 | 89 | 201 | Structureand functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q736M3 | 8.64e-20 | 198 | 317 | 207 | 321 | Gamma-D-glutamyl-L-lysine dipeptidyl-peptidase OS=Bacillus cereus (strain ATCC 10987 / NRS 248) OX=222523 GN=ykfC PE=1 SV=1 |
| Q01837 | 1.35e-18 | 206 | 312 | 413 | 509 | Probable endopeptidase p60 OS=Listeria ivanovii OX=1638 GN=iap PE=3 SV=1 |
| O64046 | 4.05e-18 | 41 | 182 | 1408 | 1537 | Probable tape measure protein OS=Bacillus phage SPbeta OX=66797 GN=yomI PE=3 SV=1 |
| O31976 | 4.05e-18 | 41 | 182 | 1408 | 1537 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
| P21171 | 7.32e-18 | 206 | 312 | 373 | 469 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
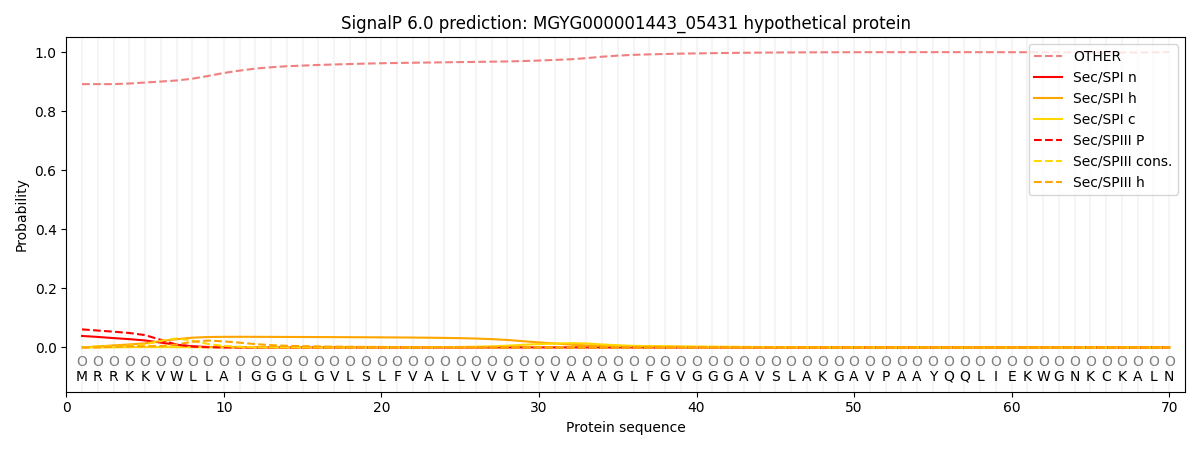
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.893666 | 0.035536 | 0.008760 | 0.000295 | 0.000154 | 0.061603 |