You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_06134
You are here: Home > Sequence: MGYG000001443_06134
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_06134 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 208962; End: 210365 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM12 | 423 | 455 | 9e-16 | 0.9411764705882353 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21112 | alphaLP-like | 8.33e-66 | 218 | 400 | 1 | 188 | alpha-lytic protease (alpha-LP), a bacterial serine protease of the chymotrypsin family, and similar proteins. This family represents the catalytic domain of alpha-lytic protease (alpha-LP) and its closely-related homologs. Alpha-lytic protease (EC 3.4.21.12; also called alpha-lytic endopeptidase), originally isolated from the myxobacterium Lysobacter enzymogenes, belongs to the MEROPS peptidase family S1, subfamily S1E (streptogrisin A subfamily). It is synthesized as a pro-enzyme, thus having two domains; the N-terminal pro-domain acts as a foldase, required transiently for the correct folding of the protease domain, and also acts as a potent inhibitor of the mature enzyme, while the C-terminal domain catalyzes the cleavage of peptide bonds. Members of the alpha-lytic protease subfamily include Nocardiopsis alba protease (NAPase), a secreted chymotrypsin from the alkaliphile Cellulomonas bogoriensis, streptogrisins (SPG-A, SPG-B, SPG-C, and SPG-D), and Thermobifida fusca protease A (TFPA). These serine proteases have characteristic kinetic stability, exhibited by their extremely slow unfolding kinetics. The active site, characteristic of serine proteases, contains the catalytic triad consisting of serine acting as a nucleophile, aspartate as an electrophile, and histidine as a base, all required for activity. This model represents the C-terminal catalytic domain of alpha-lytic proteases. |
| cd12214 | ChiA1_BD | 2.62e-17 | 423 | 467 | 1 | 45 | chitin-binding domain of Chi A1-like proteins. This group contains proteins related to the chitin binding domain of chitinase A1 (ChiA1) of Bacillus circulans WL-12. Glycosidase ChiA1 hydrolyzes chitin and is comprised of several domains: the C-terminal chitin binding domain, an N-terminal and catalytic domain, and 2 fibronectin type III-like domains. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. Bacillus circulans WL-12 ChiA1 facilitates invasion of fungal cell walls. The ChiAi chitin binding domain is required for the specific recognition of insoluble chitin. although topologically and structurally related, ChiA1 lacks the characteristic aromatic residues of Erwinia chrysanthemi endoglucanase Z (CBD(EGZ)). |
| pfam00089 | Trypsin | 5.78e-14 | 220 | 396 | 3 | 216 | Trypsin. |
| pfam02983 | Pro_Al_protease | 3.78e-06 | 145 | 200 | 1 | 57 | Alpha-lytic protease prodomain. |
| smart00495 | ChtBD3 | 4.75e-05 | 423 | 464 | 3 | 41 | Chitin-binding domain type 3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QID37532.1 | 5.21e-310 | 1 | 467 | 1 | 467 |
| QHF92780.1 | 2.88e-210 | 15 | 467 | 16 | 471 |
| QNQ35710.1 | 1.07e-199 | 16 | 467 | 3 | 457 |
| BAG20889.1 | 1.23e-198 | 18 | 467 | 5 | 457 |
| APS20807.1 | 1.63e-198 | 18 | 467 | 5 | 455 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EA3_A | 9.50e-74 | 217 | 409 | 2 | 186 | CrystalStructure Of Cellulomonas Bogoriensis Chymotrypsin [Cellulomonas bogoriensis] |
| 2PFE_A | 5.03e-66 | 216 | 405 | 1 | 185 | ChainA, Alkaline serine protease [Thermobifida fusca YX],2PFE_B Chain B, Alkaline serine protease [Thermobifida fusca YX] |
| 2OUA_A | 6.54e-64 | 216 | 405 | 1 | 188 | CrystalStructure of Nocardiopsis Protease (NAPase) [Nocardiopsis alba],2OUA_B Crystal Structure of Nocardiopsis Protease (NAPase) [Nocardiopsis alba] |
| 3M7T_A | 3.70e-44 | 216 | 405 | 1 | 197 | ChainA, Alpha-lytic protease [Lysobacter enzymogenes] |
| 1GBJ_A | 1.98e-43 | 216 | 405 | 1 | 197 | ChainA, ALPHA-LYTIC PROTEASE [Lysobacter enzymogenes],1GBK_A Alpha-lytic Protease With Met 190 Replaced By Ala Complex With Methoxysuccinyl-ala-ala-pro-alanine Boronic Acid [Lysobacter enzymogenes],1GBL_A Chain A, ALPHA-LYTIC PROTEASE [Lysobacter enzymogenes],1GBM_A Chain A, ALPHA-LYTIC PROTEASE [Lysobacter enzymogenes],2LPR_A Structural Basis For Broad Specificity In Alpha-Lytic Protease Mutants [Lysobacter enzymogenes],3LPR_A Structural Basis For Broad Specificity In Alpha-Lytic Protease Mutants [Lysobacter enzymogenes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P52320 | 2.33e-197 | 18 | 467 | 5 | 457 | Streptogrisin-C OS=Streptomyces griseus OX=1911 GN=sprC PE=3 SV=1 |
| P00778 | 8.13e-56 | 63 | 405 | 39 | 396 | Alpha-lytic protease OS=Lysobacter enzymogenes OX=69 GN=alpha-LP PE=1 SV=3 |
| P52321 | 5.43e-44 | 80 | 403 | 73 | 391 | Streptogrisin-D OS=Streptomyces griseus OX=1911 GN=sprD PE=1 SV=1 |
| P00777 | 2.31e-43 | 150 | 403 | 42 | 298 | Streptogrisin-B OS=Streptomyces griseus OX=1911 GN=sprB PE=1 SV=2 |
| Q05308 | 7.85e-41 | 65 | 406 | 63 | 397 | Serine protease 1 OS=Rarobacter faecitabidus OX=13243 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
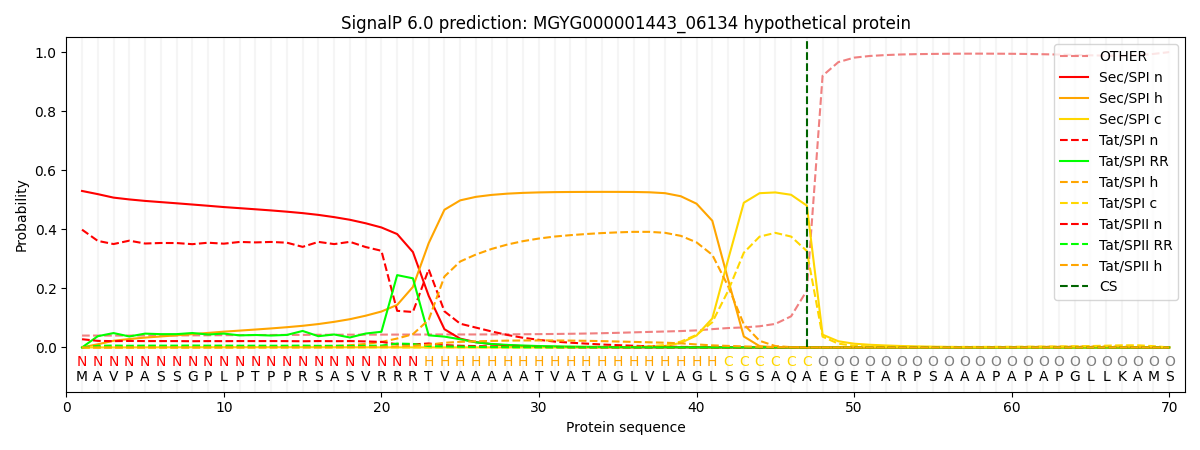
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.049339 | 0.508985 | 0.005239 | 0.406088 | 0.029669 | 0.000664 |
