You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001445_01847
Basic Information
help
| Species |
Cedecea davisae
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Cedecea; Cedecea davisae
|
| CAZyme ID |
MGYG000001445_01847
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 392 |
|
44855.72 |
7.8403 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001445 |
4852695 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1954513;
End: 1955691
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
40 |
353 |
6.7e-156 |
0.9904761904761905 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
2.79e-18 |
29 |
341 |
13 |
267 |
Cellulase (glycosyl hydrolase family 5). |
| COG2730
|
BglC |
2.22e-04 |
41 |
354 |
74 |
374 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6KDD_A
|
7.65e-13 |
43 |
342 |
44 |
295 |
endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
A1CRV0
|
2.69e-06 |
42 |
253 |
97 |
276 |
Probable glucan 1,3-beta-glucosidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=exgA PE=3 SV=2 |
|
W8QRE4
|
3.94e-06 |
101 |
350 |
124 |
345 |
Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
This protein is predicted as SP
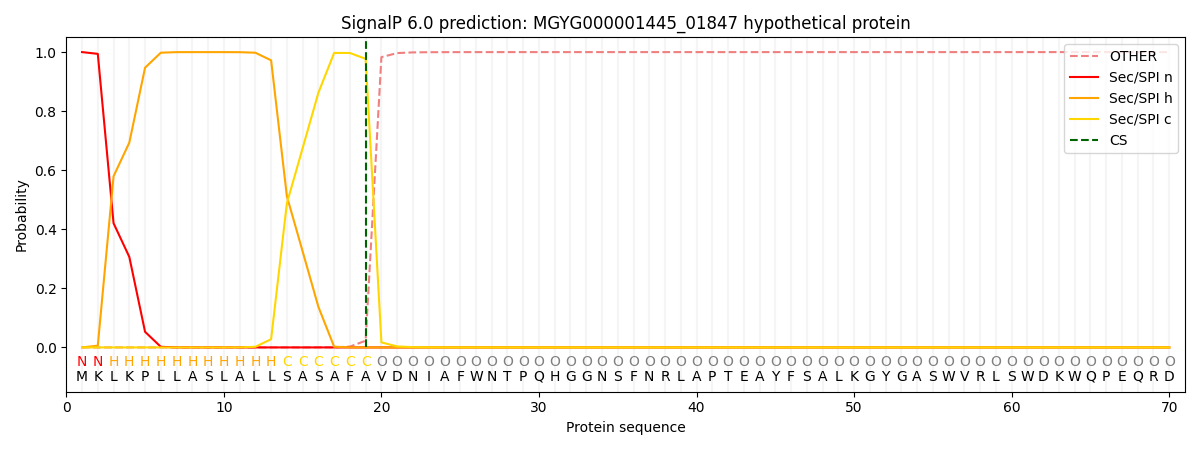
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000299
|
0.998989
|
0.000182
|
0.000179
|
0.000168
|
0.000152
|
There is no transmembrane helices in MGYG000001445_01847.

