You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001445_01909
You are here: Home > Sequence: MGYG000001445_01909
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cedecea davisae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Cedecea; Cedecea davisae | |||||||||||
| CAZyme ID | MGYG000001445_01909 | |||||||||||
| CAZy Family | GH46 | |||||||||||
| CAZyme Description | Chitosanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2040983; End: 2041459 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH46 | 26 | 139 | 3.1e-41 | 0.5045045045045045 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01374 | Glyco_hydro_46 | 1.86e-56 | 36 | 139 | 1 | 102 | Glycosyl hydrolase family 46. This family are chitosanase enzymes. |
| cd00978 | chitosanase_GH46 | 2.77e-55 | 28 | 139 | 1 | 112 | chitosanase belonging to the glycosyl hydrolase 46 family. This family is composed of the chitosanase enzymes which hydrolyzes chitosan, a biopolymer of beta (1,4)-linked-D-glucosamine (GlcN) residues produced by partial or full deacetylation of chitin. Chitosanases play a role in defense against pathogens such as fungi and are found in microorganisms, fungi, viruses, and plants. Microbial chitosanases can be divided into 3 subclasses based on the specificity of the cleavage positions for partial acetylated chitosan. Subclass I chitosanases such as N174 can split GlcN-GlcN and GlcNAc-GlcN linkages, whereas subclass II chitosanases such as Bacillus sp. no. 7-M can cleave only GlcN-GlcN linkages. Subclass III chitosanases such as MH-K1 chitosanase are the most versatile and can split both GlcN-GlcN and GlcN-GlcNAc linkages. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX97916.1 | 7.65e-91 | 1 | 139 | 1 | 139 |
| AIR03180.1 | 2.27e-63 | 1 | 139 | 2 | 139 |
| ATF92705.1 | 7.42e-62 | 1 | 139 | 2 | 139 |
| AIR62212.1 | 1.21e-60 | 7 | 139 | 7 | 139 |
| AJZ89648.1 | 2.42e-60 | 1 | 139 | 1 | 139 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ILY_A | 1.34e-45 | 20 | 151 | 13 | 146 | Abundantlysecreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4ILY_B Abundantly secreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 1CHK_A | 8.96e-41 | 20 | 145 | 3 | 127 | StreptomycesN174 Chitosanase Ph5.5 298k [Streptomyces sp. N174],1CHK_B Streptomyces N174 Chitosanase Ph5.5 298k [Streptomyces sp. N174] |
| 4OLT_A | 1.04e-38 | 20 | 145 | 11 | 138 | Chitosanasecomplex structure [Pseudomonas sp. LL2(2010)],4OLT_B Chitosanase complex structure [Pseudomonas sp. LL2(2010)] |
| 4QWP_A | 1.64e-37 | 20 | 145 | 11 | 138 | co-crystalstructure of chitosanase OU01 with substrate [Pseudomonas sp. A-01],4QWP_B co-crystal structure of chitosanase OU01 with substrate [Pseudomonas sp. A-01] |
| 7C6C_A | 2.53e-28 | 26 | 140 | 3 | 114 | ChainA, Chitosanase [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48846 | 1.72e-40 | 13 | 145 | 37 | 168 | Chitosanase OS=Nocardioides sp. (strain N106) OX=47919 GN=csn PE=3 SV=1 |
| P33665 | 6.83e-40 | 15 | 145 | 38 | 167 | Chitosanase OS=Streptomyces sp. (strain N174) OX=69019 GN=csn PE=1 SV=1 |
| O07921 | 2.86e-27 | 26 | 140 | 38 | 149 | Chitosanase OS=Bacillus subtilis (strain 168) OX=224308 GN=csn PE=1 SV=1 |
| P33673 | 1.46e-07 | 1 | 107 | 24 | 148 | Chitosanase OS=Niallia circulans OX=1397 GN=csn PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
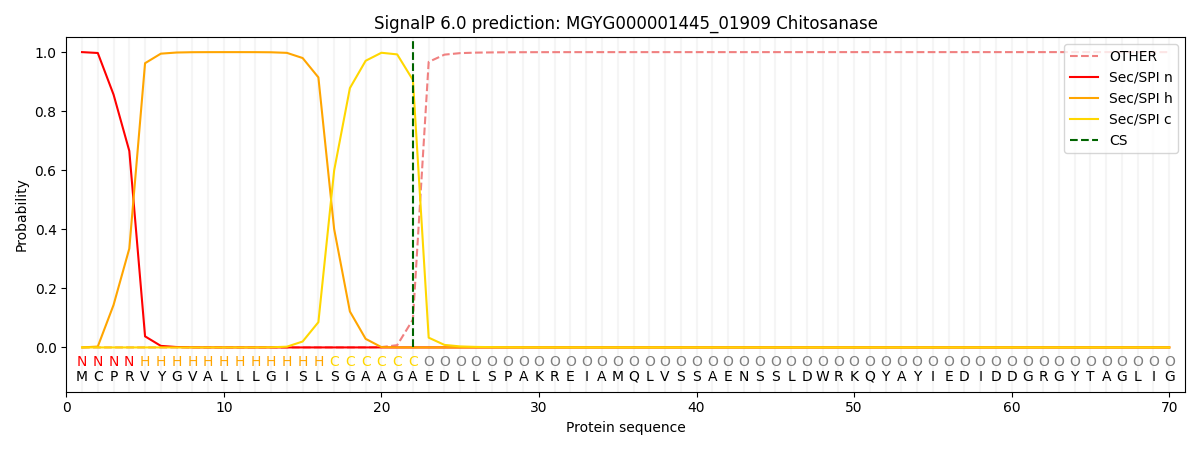
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000659 | 0.998477 | 0.000237 | 0.000216 | 0.000188 | 0.000170 |
