You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001447_01516
You are here: Home > Sequence: MGYG000001447_01516
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella oralis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella oralis | |||||||||||
| CAZyme ID | MGYG000001447_01516 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 498192; End: 499562 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06542 | GH18_EndoS-like | 9.35e-48 | 213 | 430 | 26 | 244 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
| pfam08522 | DUF1735 | 1.25e-13 | 44 | 156 | 2 | 116 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00704 | Glyco_hydro_18 | 1.91e-04 | 218 | 309 | 26 | 114 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB75661.1 | 3.55e-213 | 5 | 456 | 2 | 452 |
| QUB74142.1 | 1.01e-212 | 5 | 456 | 2 | 452 |
| AKU69596.1 | 1.13e-212 | 1 | 456 | 1 | 455 |
| EFC70999.1 | 1.60e-212 | 2 | 456 | 4 | 455 |
| QUB87240.1 | 1.18e-211 | 5 | 456 | 2 | 452 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EBN_A | 1.97e-98 | 185 | 456 | 9 | 289 | CRYSTALSTRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F1, AN ALPHA(SLASH)BETA-BARREL ENZYME ADAPTED FOR A COMPLEX SUBSTRATE [Elizabethkingia meningoseptica] |
| 6T8I_A | 1.51e-72 | 27 | 446 | 2 | 430 | Crystalstructure of wild type EndoBT-3987 from Bacteroides thetaiotamicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],6T8K_A Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 [Bacteroides thetaiotaomicron VPI-5482],6T8K_B Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 [Bacteroides thetaiotaomicron VPI-5482],6T8L_A Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man9GlcNAc product in P212121 [Bacteroides thetaiotaomicron VPI-5482],6TCW_A Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product [Bacteroides thetaiotaomicron VPI-5482],7NWF_A Chain A, Endo-beta-N-acetylglucosaminidase F1 [Bacteroides thetaiotaomicron VPI-5482] |
| 6TCV_B | 3.47e-70 | 27 | 446 | 2 | 430 | Crystalstructure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate [Bacteroides thetaiotaomicron VPI-5482] |
| 3POH_A | 2.67e-69 | 27 | 446 | 2 | 430 | Crystalstructure of an endo-beta-N-acetylglucosaminidase (BT_3987) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.55 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 1EDT_A | 2.57e-32 | 183 | 448 | 6 | 269 | ChainA, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ENDO H [Streptomyces plicatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36911 | 5.57e-97 | 185 | 456 | 59 | 339 | Endo-beta-N-acetylglucosaminidase F1 OS=Elizabethkingia meningoseptica OX=238 GN=endOF1 PE=1 SV=1 |
| P04067 | 3.55e-31 | 183 | 448 | 48 | 311 | Endo-beta-N-acetylglucosaminidase H OS=Streptomyces plicatus OX=1922 PE=1 SV=1 |
| P80036 | 4.52e-29 | 183 | 444 | 52 | 309 | Endo-beta-N-acetylglucosaminidase OS=Flavobacterium sp. (strain SK1022) OX=148444 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
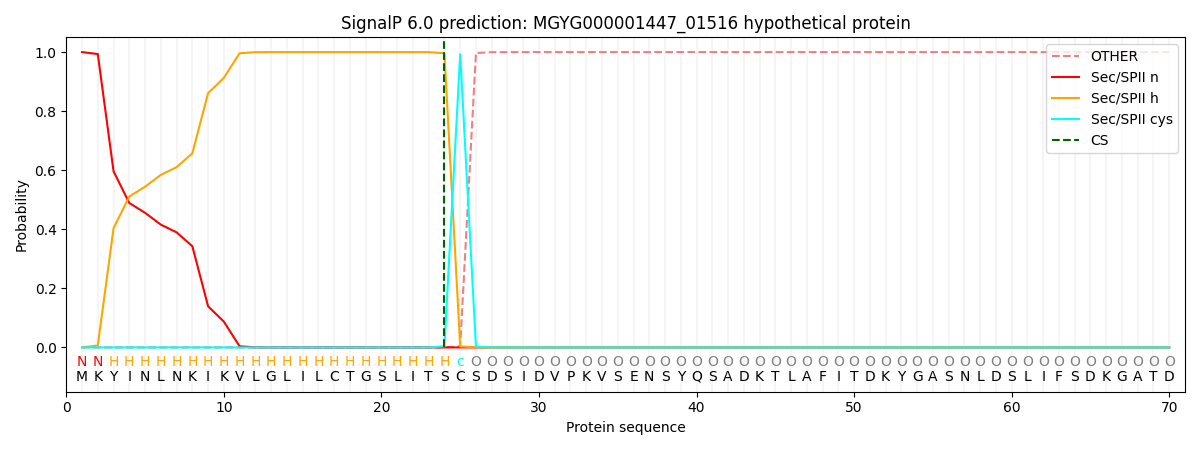
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000056 | 0.000000 | 0.000000 | 0.000000 |
