You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001448_00456
You are here: Home > Sequence: MGYG000001448_00456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
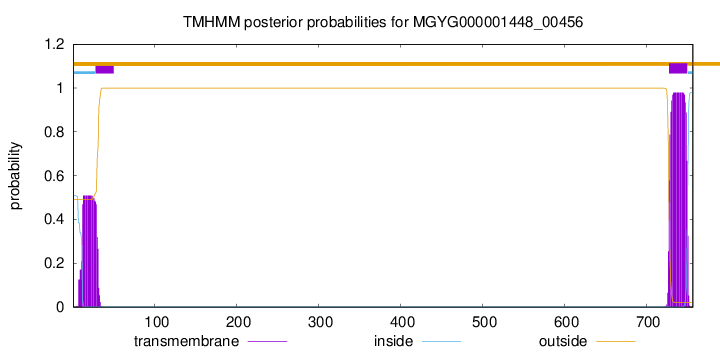
TMHMM annotations
Basic Information help
| Species | Dermabacter hominis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Dermabacteraceae; Dermabacter; Dermabacter hominis | |||||||||||
| CAZyme ID | MGYG000001448_00456 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 470144; End: 472417 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 52 | 389 | 7.9e-89 | 0.9473684210526315 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.74e-98 | 51 | 395 | 1 | 337 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 2.57e-37 | 56 | 366 | 267 | 620 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 1.12e-25 | 75 | 379 | 1 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13859 | BNR_3 | 4.60e-12 | 74 | 294 | 8 | 213 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam10633 | NPCBM_assoc | 9.89e-07 | 414 | 477 | 3 | 68 | NPCBM-associated, NEW3 domain of alpha-galactosidase. The English-language version of the first reference can be found on pages 388-399 of the above. This domain has been named NEW3 but its actual function is not known. It is found on proteins which are bacterial galactosidases. The domain is associated with the NPCBM family, pfam08305, a novel putative carbohydrate binding module found at the N-terminus of glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANP28544.1 | 0.0 | 1 | 754 | 2 | 752 |
| ATH95829.1 | 0.0 | 1 | 509 | 2 | 510 |
| AZQ76181.1 | 4.47e-232 | 2 | 625 | 5 | 622 |
| ALE04399.1 | 3.83e-156 | 26 | 626 | 27 | 674 |
| AIY02020.1 | 3.04e-155 | 26 | 625 | 27 | 671 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1EUR_A | 8.08e-114 | 42 | 403 | 3 | 364 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 1EUT_A | 1.43e-113 | 42 | 494 | 3 | 450 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1WCQ_A | 2.72e-112 | 48 | 494 | 5 | 446 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BZD_A | 5.36e-112 | 48 | 494 | 5 | 446 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 1W8N_A | 7.53e-112 | 48 | 494 | 5 | 446 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.80e-112 | 37 | 494 | 40 | 492 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P31206 | 2.92e-25 | 64 | 384 | 203 | 528 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
| P29767 | 1.72e-21 | 57 | 545 | 388 | 968 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| Q99519 | 6.21e-17 | 59 | 386 | 72 | 398 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
| Q5RAF4 | 6.21e-17 | 59 | 386 | 72 | 398 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
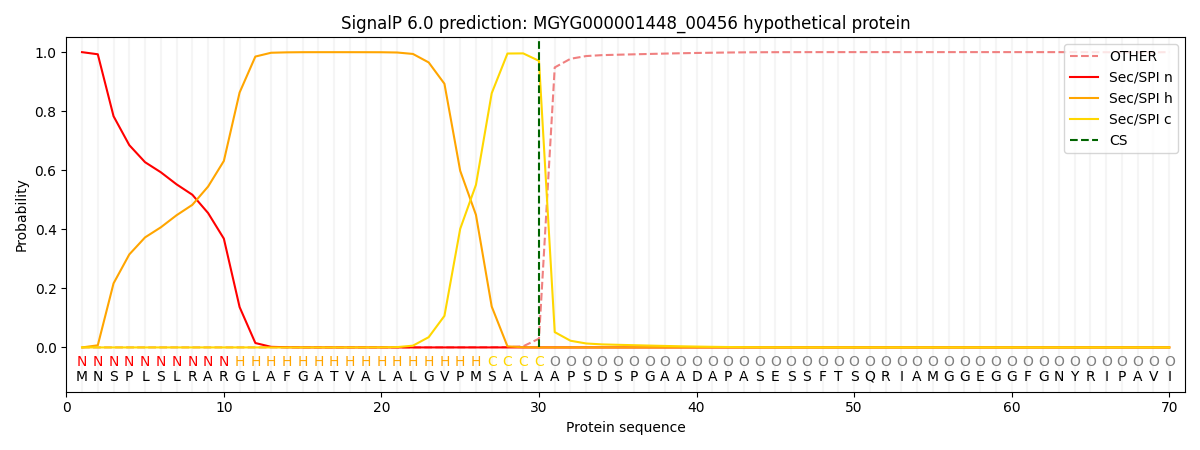
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001005 | 0.997572 | 0.000304 | 0.000539 | 0.000332 | 0.000236 |