You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001449_01295
You are here: Home > Sequence: MGYG000001449_01295
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
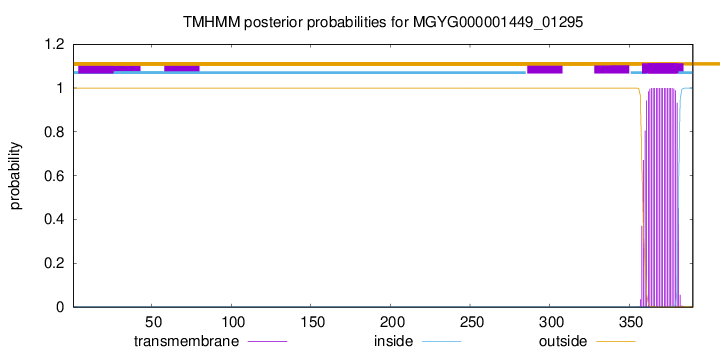
TMHMM annotations
Basic Information help
| Species | Nosocomiicoccus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Salinicoccaceae; Nosocomiicoccus; Nosocomiicoccus massiliensis | |||||||||||
| CAZyme ID | MGYG000001449_01295 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | 1,4-alpha-glucan branching enzyme GlgB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20022; End: 21194 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH13 | 1 | 238 | 6.7e-21 | 0.8896321070234113 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11313 | AmyAc_arch_bac_AmyA | 2.96e-15 | 1 | 196 | 34 | 255 | Alpha amylase catalytic domain found in archaeal and bacterial Alpha-amylases (also called 1,4-alpha-D-glucan-4-glucanohydrolase). AmyA (EC 3.2.1.1) catalyzes the hydrolysis of alpha-(1,4) glycosidic linkages of glycogen, starch, related polysaccharides, and some oligosaccharides. This group includes firmicutes, bacteroidetes, and proteobacteria. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| smart00642 | Aamy | 4.69e-13 | 1 | 58 | 31 | 90 | Alpha-amylase domain. |
| cd00551 | AmyAc_family | 1.61e-12 | 1 | 59 | 37 | 97 | Alpha amylase catalytic domain family. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; and C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost this catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| pfam00128 | Alpha-amylase | 4.44e-12 | 1 | 58 | 16 | 72 | Alpha amylase, catalytic domain. Alpha amylase is classified as family 13 of the glycosyl hydrolases. The structure is an 8 stranded alpha/beta barrel containing the active site, interrupted by a ~70 a.a. calcium-binding domain protruding between beta strand 3 and alpha helix 3, and a carboxyl-terminal Greek key beta-barrel domain. |
| COG0366 | AmyA | 1.11e-11 | 1 | 60 | 41 | 99 | Glycosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYA47208.1 | 7.15e-230 | 1 | 390 | 78 | 467 |
| QYA48833.1 | 1.67e-228 | 1 | 390 | 78 | 467 |
| QQD85930.1 | 5.36e-128 | 1 | 389 | 78 | 467 |
| AKG73328.1 | 6.97e-113 | 1 | 389 | 78 | 471 |
| QQB05469.1 | 7.80e-105 | 1 | 389 | 76 | 462 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5A2B_A | 1.73e-30 | 1 | 359 | 90 | 489 | CrystalStructure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass [Anoxybacillus ayderensis],5A2C_A Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass [Anoxybacillus ayderensis] |
| 5A2A_A | 1.00e-29 | 1 | 358 | 56 | 454 | CrystalStructure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass [Anoxybacillus ayderensis] |
| 4E2O_A | 1.90e-24 | 1 | 354 | 57 | 452 | Crystalstructure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose [Geobacillus thermoleovorans CCB_US3_UF5] |
| 1UOK_A | 5.87e-07 | 1 | 65 | 43 | 106 | CrystalStructure Of B. Cereus Oligo-1,6-Glucosidase [Bacillus cereus] |
| 5DO8_A | 1.80e-06 | 1 | 65 | 44 | 107 | 1.8Angstrom crystal structure of Listeria monocytogenes Lmo0184 alpha-1,6-glucosidase [Listeria monocytogenes EGD-e],5DO8_B 1.8 Angstrom crystal structure of Listeria monocytogenes Lmo0184 alpha-1,6-glucosidase [Listeria monocytogenes EGD-e],5DO8_C 1.8 Angstrom crystal structure of Listeria monocytogenes Lmo0184 alpha-1,6-glucosidase [Listeria monocytogenes EGD-e] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P43473 | 2.43e-06 | 1 | 65 | 45 | 108 | Alpha-glucosidase OS=Pediococcus pentosaceus OX=1255 GN=agl PE=3 SV=1 |
| P21332 | 3.22e-06 | 1 | 65 | 43 | 106 | Oligo-1,6-glucosidase OS=Bacillus cereus OX=1396 GN=malL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
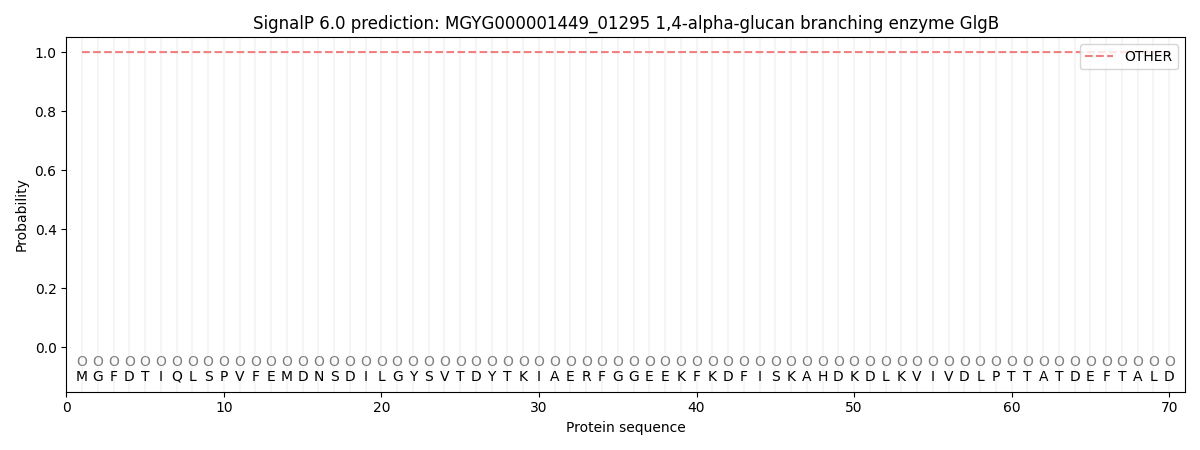
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000074 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |