You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001451_00164
You are here: Home > Sequence: MGYG000001451_00164
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
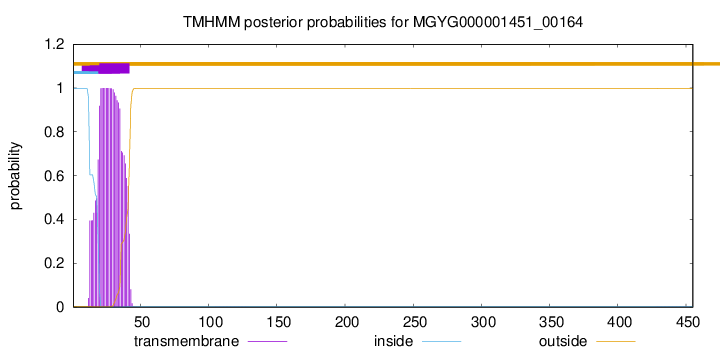
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A antibioticophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A antibioticophila | |||||||||||
| CAZyme ID | MGYG000001451_00164 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 177528; End: 178895 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 2.07e-18 | 106 | 360 | 27 | 247 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 1.23e-11 | 110 | 453 | 52 | 355 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PRK11097 | PRK11097 | 3.19e-07 | 106 | 305 | 47 | 234 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDM47530.1 | 2.01e-231 | 47 | 452 | 38 | 444 |
| AIQ25865.1 | 6.31e-170 | 47 | 452 | 978 | 1379 |
| BBI36306.1 | 7.94e-169 | 47 | 455 | 51 | 443 |
| QJD86164.1 | 5.44e-159 | 46 | 452 | 60 | 466 |
| QAV17253.1 | 7.40e-153 | 40 | 451 | 48 | 448 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XD0_A | 1.04e-136 | 10 | 450 | 3 | 408 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 7CJU_A | 1.01e-85 | 42 | 451 | 6 | 390 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 1V5C_A | 1.06e-85 | 45 | 451 | 3 | 384 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 1CEM_A | 3.07e-41 | 48 | 455 | 4 | 363 | ChainA, CELLULASE CELA (1,4-BETA-D-GLUCAN-GLUCANOHYDROLASE) [Acetivibrio thermocellus],1IS9_A Chain A, endoglucanase A [Acetivibrio thermocellus] |
| 1KWF_A | 8.21e-40 | 48 | 455 | 4 | 363 | ChainA, Endoglucanase A [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19254 | 1.62e-135 | 7 | 450 | 2 | 408 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P29019 | 1.99e-80 | 45 | 451 | 59 | 440 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
| A3DC29 | 2.74e-39 | 11 | 455 | 5 | 395 | Endoglucanase A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celA PE=1 SV=1 |
| P37701 | 3.15e-31 | 48 | 447 | 37 | 388 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
| P37699 | 4.69e-29 | 24 | 447 | 14 | 388 | Endoglucanase C OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCC PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
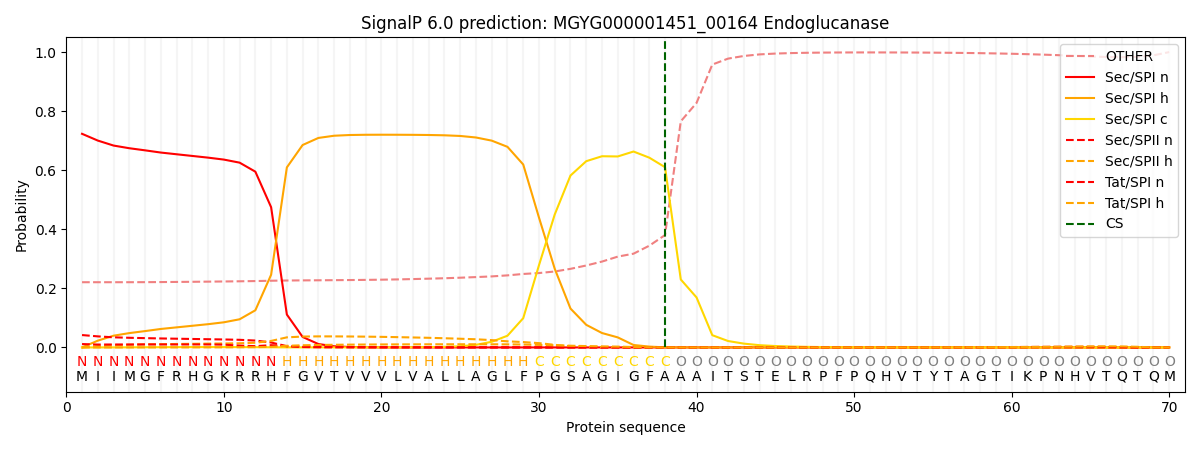
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.237828 | 0.691955 | 0.045495 | 0.018217 | 0.004640 | 0.001849 |