You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001451_00486
You are here: Home > Sequence: MGYG000001451_00486
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
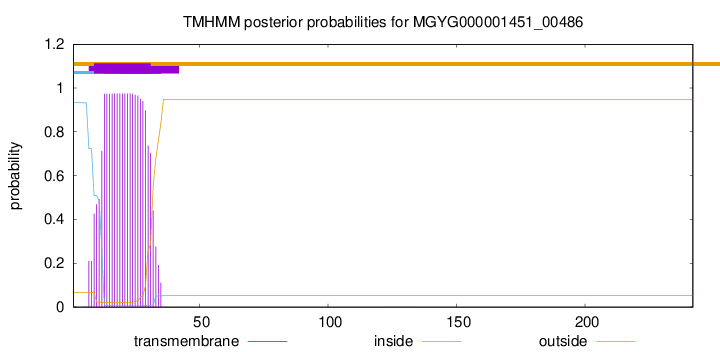
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A antibioticophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A antibioticophila | |||||||||||
| CAZyme ID | MGYG000001451_00486 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 552418; End: 553146 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 68 | 222 | 2.9e-61 | 0.5507246376811594 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.71e-41 | 61 | 235 | 1 | 177 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.83e-31 | 29 | 192 | 29 | 199 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANF96686.1 | 5.72e-97 | 4 | 223 | 3 | 225 |
| AIQ33839.1 | 1.08e-96 | 41 | 223 | 42 | 224 |
| ALF95239.2 | 5.47e-96 | 40 | 223 | 37 | 220 |
| AIQ22098.1 | 1.21e-95 | 41 | 223 | 42 | 224 |
| ASA25821.1 | 1.71e-95 | 40 | 241 | 41 | 240 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5E0C_A | 7.32e-77 | 41 | 233 | 7 | 199 | StructuralInsight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
| 5XRC_A | 2.19e-76 | 41 | 233 | 40 | 232 | ATrimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_B A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_C A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10] |
| 4V2X_A | 9.29e-74 | 15 | 233 | 33 | 247 | ChainA, Endo-beta-1,4-glucanase (cellulase B) [Halalkalibacterium halodurans] |
| 5E09_A | 5.86e-72 | 41 | 233 | 7 | 199 | StructuralInsight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
| 4YZP_A | 8.75e-50 | 48 | 222 | 28 | 192 | Crystalstructure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis [Bacillus licheniformis],4YZT_A Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose [Bacillus licheniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23550 | 4.82e-88 | 42 | 223 | 35 | 217 | Endoglucanase B OS=Paenibacillus lautus OX=1401 GN=celB PE=3 SV=1 |
| P28621 | 1.13e-44 | 48 | 222 | 46 | 221 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P28623 | 7.75e-44 | 48 | 222 | 47 | 222 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P23660 | 2.98e-42 | 48 | 222 | 31 | 211 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P23661 | 3.78e-42 | 38 | 222 | 65 | 253 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
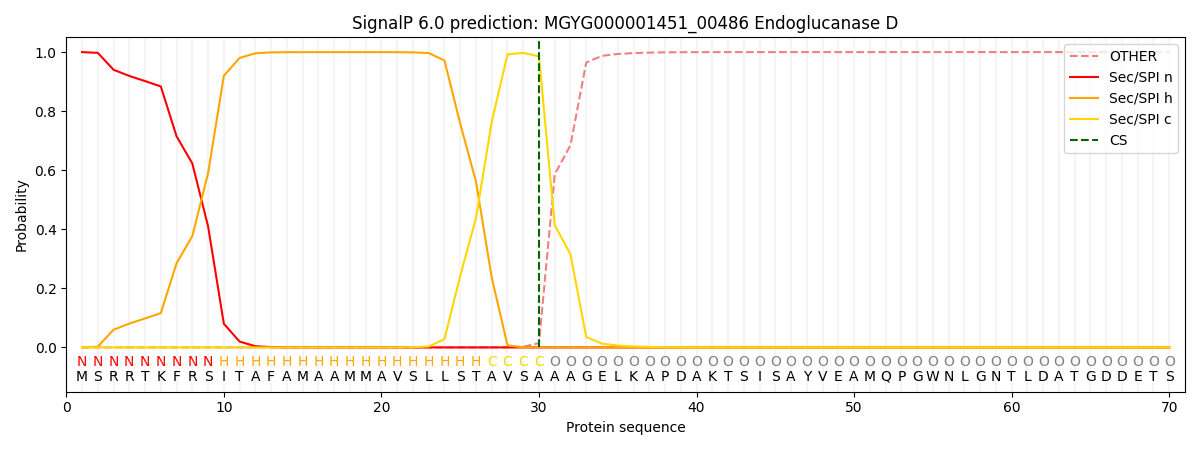
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000363 | 0.998900 | 0.000185 | 0.000212 | 0.000174 | 0.000157 |