You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001451_05011
You are here: Home > Sequence: MGYG000001451_05011
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
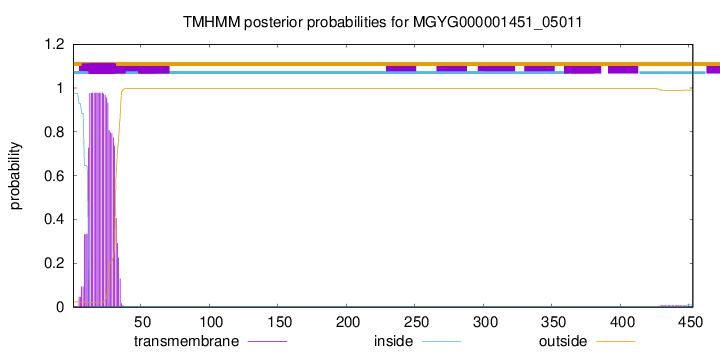
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A antibioticophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A antibioticophila | |||||||||||
| CAZyme ID | MGYG000001451_05011 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67742; End: 69103 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM16 | 34 | 154 | 1.4e-16 | 0.9827586206896551 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033675 | NTTRR-F1 | 2.20e-05 | 33 | 173 | 3 | 155 | NTTRR-F1 domain. NTTRR-F1 (N-terminal To Repetitive Region - Firmicutes 1) is a homology domain found strictly as the N-terminal non-repetitive region of otherwise highly repetitive proteins of various Firmicutes. The repetitive region that follows typically is collagen-like, with every third residue a glycine. |
| pfam02018 | CBM_4_9 | 1.07e-04 | 34 | 154 | 2 | 129 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATB41231.1 | 3.74e-204 | 19 | 453 | 18 | 449 |
| QRO01845.1 | 3.53e-202 | 19 | 453 | 18 | 449 |
| QAV19451.1 | 7.63e-113 | 32 | 449 | 59 | 480 |
| AVM43010.1 | 1.83e-20 | 32 | 176 | 268 | 433 |
| AUS96050.1 | 4.79e-15 | 34 | 172 | 816 | 953 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZG90 | 1.71e-09 | 35 | 172 | 291 | 421 | Keratan-sulfate endo-1,4-beta-galactosidase OS=Sphingobacterium multivorum OX=28454 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000314 | 0.998963 | 0.000205 | 0.000183 | 0.000161 | 0.000144 |