You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001453_04772
Basic Information
help
| Species |
Aneurinibacillus aneurinilyticus
|
| Lineage |
Bacteria; Firmicutes; Bacilli; Aneurinibacillales; Aneurinibacillaceae; Aneurinibacillus; Aneurinibacillus aneurinilyticus
|
| CAZyme ID |
MGYG000001453_04772
|
| CAZy Family |
GT89 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 639 |
|
71923.12 |
9.7003 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001453 |
5302959 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 9770;
End: 11689
Strand: +
|
No EC number prediction in MGYG000001453_04772.
| Family |
Start |
End |
Evalue |
family coverage |
| GT89 |
17 |
387 |
2.8e-55 |
0.670863309352518 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
0.002 |
17 |
368 |
13 |
354 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
0.006 |
70 |
219 |
4 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O53582
|
2.94e-18 |
17 |
464 |
4 |
535 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=aftB PE=1 SV=3 |
|
Q8NLR0
|
3.32e-11 |
35 |
346 |
72 |
419 |
Terminal beta-(1->2)-arabinofuranosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=aftB PE=3 SV=1 |
This protein is predicted as OTHER
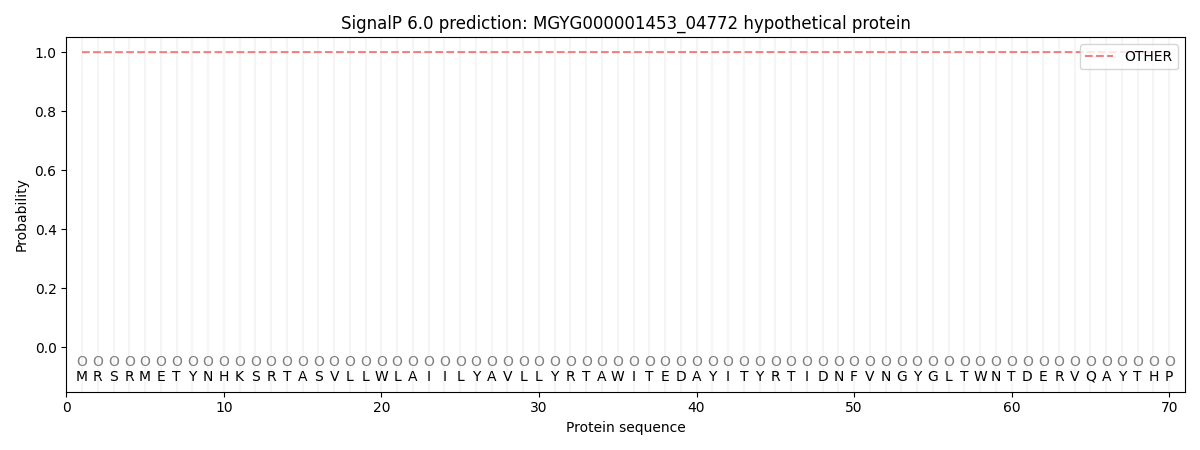
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999820
|
0.000132
|
0.000006
|
0.000001
|
0.000000
|
0.000084
|
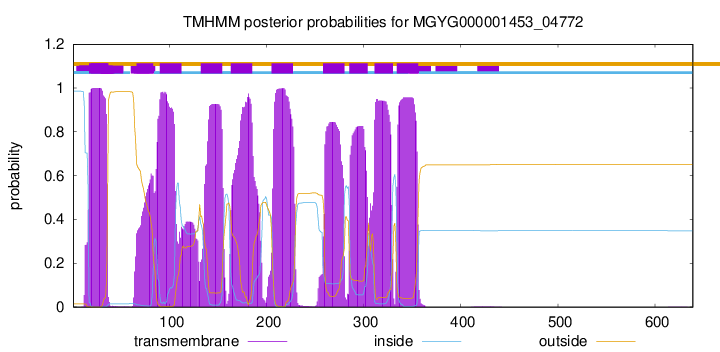
| start |
end |
| 17 |
36 |
| 66 |
83 |
| 90 |
112 |
| 132 |
154 |
| 163 |
185 |
| 205 |
227 |
| 258 |
280 |
| 285 |
304 |
| 311 |
330 |
| 334 |
356 |


