You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001457_00566
You are here: Home > Sequence: MGYG000001457_00566
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas_B luteola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_B; Pseudomonas_B luteola | |||||||||||
| CAZyme ID | MGYG000001457_00566 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 545121; End: 549494 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 134 | 303 | 3.7e-36 | 0.9823529411764705 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03030 | CelA | 0.0 | 13 | 688 | 13 | 710 | cellulose synthase catalytic subunit (UDP-forming). Cellulose synthase catalyzes the beta-1,4 polymerization of glucose residues in the formation of cellulose. In bacteria, the substrate is UDP-glucose. The synthase consists of two subunits (or domains in the frequent cases where it is encoded as a single polypeptide), the catalytic domain modelled here and the regulatory domain (pfam03170). The regulatory domain binds the allosteric activator cyclic di-GMP. The protein is membrane-associated and probably assembles into multimers such that the individual cellulose strands can self-assemble into multi-strand fibrils. |
| PRK11114 | PRK11114 | 0.0 | 741 | 1454 | 45 | 755 | cellulose biosynthesis cyclic di-GMP-binding regulatory protein BcsB. |
| PRK11498 | bcsA | 0.0 | 1 | 677 | 130 | 807 | cellulose synthase catalytic subunit; Provisional |
| pfam03170 | BcsB | 0.0 | 753 | 1358 | 1 | 605 | Bacterial cellulose synthase subunit. This family includes bacterial proteins involved in cellulose synthesis. Cellulose synthesis has been identified in several bacteria. In Agrobacterium tumefaciens, for instance, cellulose has a pathogenic role: it allows the bacteria to bind tightly to their host plant cells. While several enzymatic steps are involved in cellulose synthesis, potentially the only step unique to this pathway is that catalyzed by cellulose synthase. This enzyme is a multi subunit complex. This family encodes a subunit that is thought to bind the positive effector cyclic di-GMP. This subunit is found in several different bacterial cellulose synthase enzymes. The first recognized sequence for this subunit is BcsB. In the AcsII cellulose synthase, this subunit and the subunit corresponding to BcsA are found in the same protein. Indeed, this alignment only includes the C-terminal half of the AcsAII synthase, which corresponds to BcsB. |
| cd06421 | CESA_CelA_like | 5.08e-108 | 131 | 363 | 1 | 234 | CESA_CelA_like are involved in the elongation of the glucan chain of cellulose. Family of proteins related to Agrobacterium tumefaciens CelA and Gluconacetobacter xylinus BscA. These proteins are involved in the elongation of the glucan chain of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues. They are putative catalytic subunit of cellulose synthase, which is a glycosyltransferase using UDP-glucose as the substrate. The catalytic subunit is an integral membrane protein with 6 transmembrane segments and it is postulated that the protein is anchored in the membrane at the N-terminal end. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEU28406.1 | 0.0 | 1 | 1457 | 1 | 1457 |
| AYN94005.1 | 0.0 | 1 | 1457 | 1 | 1457 |
| QXY92926.1 | 0.0 | 17 | 1452 | 20 | 1456 |
| QQE90988.1 | 0.0 | 3 | 1453 | 2 | 1451 |
| AJE22983.1 | 0.0 | 3 | 1453 | 8 | 1457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LBY_A | 1.60e-202 | 46 | 676 | 188 | 819 | ChainA, Cellulose synthase catalytic subunit [UDP-forming] [Escherichia coli K-12] |
| 5EJ1_A | 2.52e-104 | 70 | 591 | 67 | 605 | ChainA, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
| 4HG6_A | 1.52e-103 | 70 | 591 | 79 | 617 | ChainA, Cellulose Synthase Subunit A [Cereibacter sphaeroides] |
| 4P00_A | 1.56e-103 | 70 | 591 | 80 | 618 | ChainA, Cellulose Synthase A subunit [Cereibacter sphaeroides 2.4.1],4P02_A Chain A, Cellulose Synthase subunit A [Cereibacter sphaeroides 2.4.1],5EIY_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1],5EJZ_A Chain A, Putative cellulose synthase [Cereibacter sphaeroides 2.4.1] |
| 6YG8_A | 2.70e-93 | 768 | 1457 | 86 | 778 | ChainA, Bacterial cellulose secretion regulator BcsB [Escherichia coli],6YG8_B Chain B, Bacterial cellulose secretion regulator BcsB [Escherichia coli],6YG8_C Chain C, Bacterial cellulose secretion regulator BcsB [Escherichia coli],6YG8_D Chain D, Bacterial cellulose secretion regulator BcsB [Escherichia coli],6YG8_E Chain E, Bacterial cellulose secretion regulator BcsB [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9WX75 | 0.0 | 38 | 1455 | 53 | 1511 | Putative cellulose synthase 3 OS=Komagataeibacter xylinus OX=28448 GN=bcsABII-B PE=3 SV=1 |
| Q9RBJ2 | 0.0 | 38 | 1455 | 53 | 1511 | Putative cellulose synthase 2 OS=Komagataeibacter xylinus OX=28448 GN=bcsABII-A PE=3 SV=1 |
| Q59167 | 5.38e-313 | 46 | 1453 | 62 | 1582 | Cellulose synthase 2 OS=Komagataeibacter hansenii OX=436 GN=acsAII PE=3 SV=1 |
| P0CW87 | 6.81e-311 | 32 | 1437 | 50 | 1529 | Cellulose synthase 1 OS=Komagataeibacter xylinus OX=28448 GN=acsAB PE=1 SV=1 |
| Q76KJ8 | 2.96e-309 | 32 | 1437 | 50 | 1529 | Cellulose synthase 1 OS=Komagataeibacter hansenii OX=436 GN=acsAB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000039 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
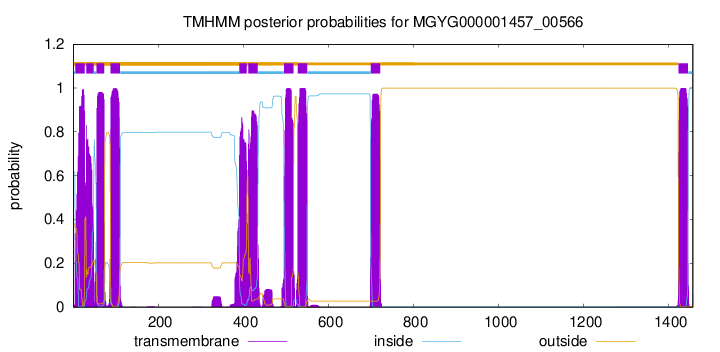
| start | end |
|---|---|
| 5 | 27 |
| 31 | 48 |
| 55 | 73 |
| 88 | 110 |
| 391 | 408 |
| 412 | 434 |
| 496 | 518 |
| 528 | 550 |
| 700 | 722 |
| 1423 | 1445 |
