You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001457_01435
You are here: Home > Sequence: MGYG000001457_01435
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas_B luteola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_B; Pseudomonas_B luteola | |||||||||||
| CAZyme ID | MGYG000001457_01435 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Pectate lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1436576; End: 1437721 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 99 | 310 | 2.8e-84 | 0.9859154929577465 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.23e-61 | 36 | 380 | 34 | 345 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 3.50e-52 | 107 | 310 | 12 | 186 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 2.62e-47 | 80 | 310 | 3 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEU27652.1 | 1.02e-277 | 1 | 381 | 1 | 381 |
| AYN93235.1 | 5.63e-275 | 1 | 381 | 1 | 381 |
| QEU27653.1 | 1.24e-197 | 16 | 381 | 16 | 379 |
| AYN93236.1 | 1.24e-197 | 16 | 381 | 16 | 379 |
| QEU27654.1 | 1.65e-193 | 16 | 381 | 17 | 380 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1PCL_A | 3.10e-92 | 33 | 379 | 5 | 355 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
| 5AMV_A | 4.82e-88 | 30 | 380 | 5 | 399 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
| 1BN8_A | 8.96e-88 | 30 | 380 | 26 | 420 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
| 2BSP_A | 2.52e-87 | 30 | 380 | 26 | 420 | ChainA, PROTEIN (PECTATE LYASE) [Bacillus subtilis] |
| 2NZM_A | 3.84e-87 | 30 | 380 | 5 | 399 | ChainA, Pectate lyase [Bacillus subtilis],2O04_A Chain A, Pectate lyase [Bacillus subtilis],2O0V_A Chain A, Pectate lyase [Bacillus subtilis],2O0W_A Chain A, Pectate lyase [Bacillus subtilis],2O17_A Chain A, Pectate lyase [Bacillus subtilis],2O1D_A Chain A, Pectate lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56806 | 2.16e-163 | 33 | 379 | 31 | 377 | Pectate lyase OS=Xanthomonas campestris pv. malvacearum OX=86040 PE=3 SV=1 |
| P72242 | 3.59e-160 | 15 | 379 | 13 | 379 | Pectate lyase OS=Pseudomonas amygdali pv. lachrymans OX=53707 GN=pelP PE=3 SV=1 |
| Q59671 | 5.27e-160 | 14 | 379 | 16 | 380 | Pectate lyase OS=Pseudomonas fluorescens OX=294 GN=pel PE=3 SV=1 |
| Q60140 | 7.48e-160 | 19 | 379 | 20 | 379 | Pectate lyase OS=Pseudomonas viridiflava OX=33069 GN=pel PE=3 SV=1 |
| Q51915 | 4.30e-159 | 25 | 379 | 23 | 380 | Pectate lyase OS=Pseudomonas marginalis OX=298 GN=pel PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
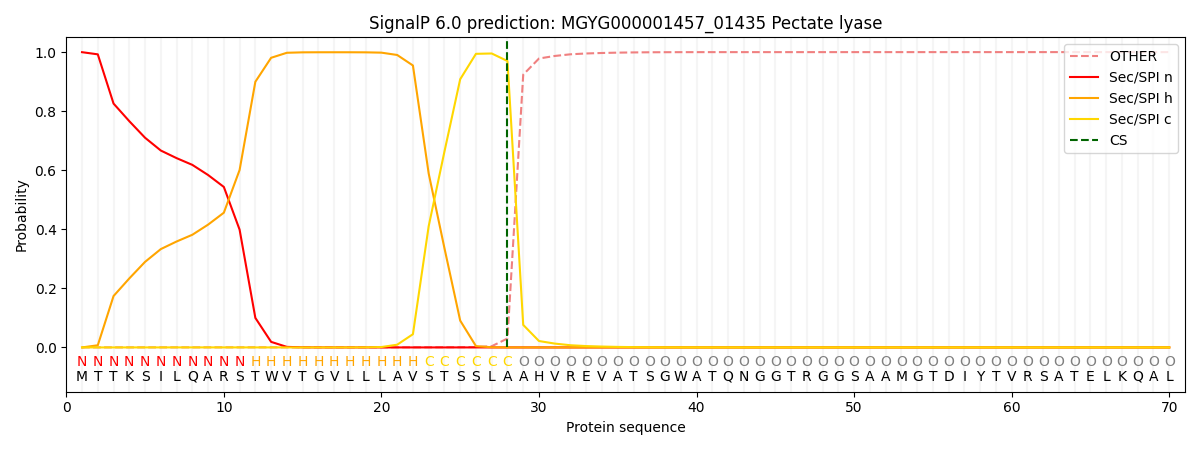
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001044 | 0.998078 | 0.000256 | 0.000212 | 0.000191 | 0.000179 |
