You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001457_04572
You are here: Home > Sequence: MGYG000001457_04572
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
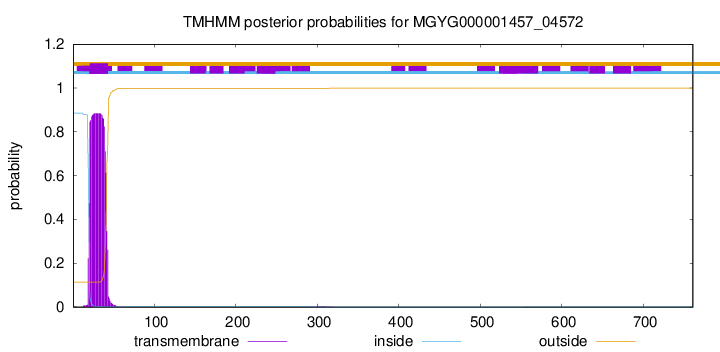
TMHMM annotations
Basic Information help
| Species | Pseudomonas_B luteola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_B; Pseudomonas_B luteola | |||||||||||
| CAZyme ID | MGYG000001457_04572 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 593661; End: 595946 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 39 | 318 | 3.9e-97 | 0.9893617021276596 |
| GH5 | 447 | 726 | 2.5e-90 | 0.9893617021276596 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 9.83e-42 | 60 | 313 | 21 | 270 | Cellulase (glycosyl hydrolase family 5). |
| pfam00150 | Cellulase | 8.99e-36 | 471 | 721 | 24 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.52e-20 | 38 | 344 | 47 | 395 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG2730 | BglC | 2.29e-14 | 431 | 752 | 36 | 395 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| NF033201 | Vip_LPXTG_Lm | 2.00e-08 | 350 | 403 | 272 | 329 | cell invasion LPXTG protein Vip. Vip (Virulence protein), like the LPXTG-type internalins, is an LPXTG-anchored surface protein of the mammalian cell-invading pathogen Listeria monocytogenes, but absent from the related species Listeria innocua. For certain cell types, Vip is required for Listeria's ability to invade. It appears to bind the endoplasmic reticulum (ER) resident chaperone Gp96 as its receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEU27537.1 | 0.0 | 16 | 761 | 1 | 749 |
| AYN96127.1 | 0.0 | 16 | 761 | 1 | 798 |
| QEU02828.1 | 2.24e-305 | 21 | 759 | 17 | 893 |
| APQ14298.1 | 1.15e-303 | 23 | 759 | 20 | 896 |
| QDD87797.1 | 2.47e-303 | 23 | 759 | 20 | 898 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EE9_A | 2.99e-127 | 35 | 350 | 1 | 317 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 8.41e-127 | 35 | 350 | 1 | 317 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 1.15e-126 | 35 | 350 | 1 | 317 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 1.19e-126 | 35 | 350 | 1 | 317 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
| 5LJF_A | 2.36e-126 | 35 | 350 | 1 | 317 | ChainA, Endoglucanase [uncultured bacterium],5LJF_B Chain B, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17974 | 2.61e-52 | 40 | 340 | 121 | 425 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| P58599 | 1.69e-47 | 448 | 749 | 119 | 424 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| B8MW97 | 8.24e-43 | 39 | 328 | 34 | 319 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
| Q2UPQ4 | 8.24e-43 | 39 | 328 | 34 | 319 | Probable endo-beta-1,4-glucanase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglB PE=3 SV=1 |
| A1DME8 | 2.87e-40 | 448 | 735 | 31 | 314 | Probable endo-beta-1,4-glucanase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
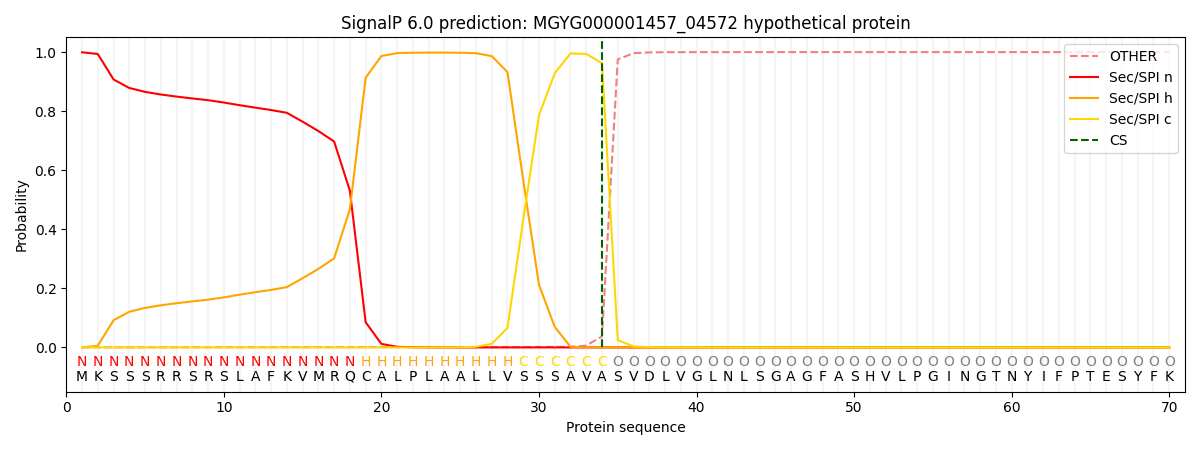
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001465 | 0.996840 | 0.001015 | 0.000232 | 0.000207 | 0.000204 |