You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001458_00629
You are here: Home > Sequence: MGYG000001458_00629
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cetobacterium_A somerae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Cetobacterium_A; Cetobacterium_A somerae | |||||||||||
| CAZyme ID | MGYG000001458_00629 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4459; End: 5637 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 57 | 350 | 3.9e-100 | 0.9896193771626297 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 5.84e-61 | 32 | 381 | 2 | 309 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 1.77e-09 | 37 | 349 | 4 | 266 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 0.003 | 30 | 294 | 49 | 286 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCK83069.1 | 4.30e-140 | 25 | 390 | 46 | 423 |
| AAC71692.1 | 4.43e-140 | 25 | 390 | 46 | 423 |
| QXJ38243.1 | 1.22e-139 | 25 | 390 | 46 | 423 |
| ACM23521.1 | 2.00e-137 | 5 | 386 | 2 | 390 |
| CAB56856.1 | 2.00e-137 | 5 | 386 | 2 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PZ9_A | 1.06e-136 | 35 | 386 | 21 | 380 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 1.93e-133 | 35 | 390 | 7 | 370 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 4QP0_A | 8.78e-72 | 26 | 392 | 4 | 359 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3WH9_A | 1.66e-64 | 26 | 372 | 2 | 334 | Theligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger],3WH9_B The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger] |
| 3ZIZ_A | 6.45e-64 | 34 | 349 | 23 | 318 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1D8Y6 | 1.23e-68 | 4 | 372 | 3 | 358 | Probable mannan endo-1,4-beta-mannosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manA PE=3 SV=1 |
| B8NVK8 | 6.95e-68 | 24 | 372 | 41 | 374 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manA PE=3 SV=2 |
| Q2TXJ2 | 1.51e-66 | 24 | 372 | 41 | 374 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manA PE=3 SV=1 |
| Q0CCD0 | 9.68e-65 | 24 | 372 | 39 | 373 | Probable mannan endo-1,4-beta-mannosidase A-1 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=manA-1 PE=3 SV=1 |
| Q00012 | 2.20e-63 | 22 | 372 | 28 | 364 | Mannan endo-1,4-beta-mannosidase A OS=Aspergillus aculeatus OX=5053 GN=manA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
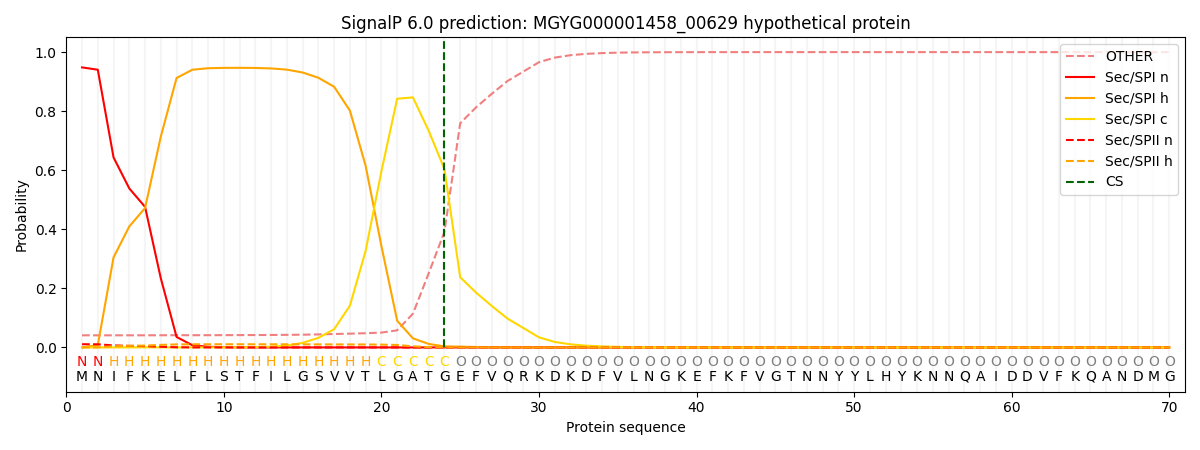
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.045331 | 0.941946 | 0.011881 | 0.000283 | 0.000252 | 0.000266 |
