You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001458_02402
You are here: Home > Sequence: MGYG000001458_02402
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cetobacterium_A somerae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Cetobacterium_A; Cetobacterium_A somerae | |||||||||||
| CAZyme ID | MGYG000001458_02402 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 46826; End: 48217 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 10 | 373 | 6.9e-63 | 0.6444444444444445 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.01e-30 | 8 | 376 | 4 | 369 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.54e-10 | 65 | 224 | 1 | 153 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 9.84e-10 | 14 | 390 | 13 | 384 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVQ28191.1 | 4.75e-110 | 4 | 457 | 2 | 486 |
| SQI99970.1 | 4.75e-110 | 4 | 457 | 2 | 486 |
| AVQ30207.1 | 2.58e-103 | 4 | 457 | 2 | 485 |
| BBA52631.1 | 2.55e-100 | 4 | 457 | 2 | 485 |
| AVQ18501.1 | 3.40e-71 | 3 | 462 | 2 | 488 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8D339 | 1.53e-12 | 20 | 362 | 23 | 362 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Wigglesworthia glossinidia brevipalpis OX=36870 GN=arnT PE=3 SV=1 |
| O67270 | 2.22e-10 | 36 | 378 | 29 | 353 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| A0KGY4 | 9.85e-10 | 14 | 373 | 10 | 360 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1 |
| Q3KCC9 | 7.23e-09 | 3 | 388 | 14 | 395 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1 |
| Q1C744 | 9.40e-09 | 1 | 330 | 1 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000006 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
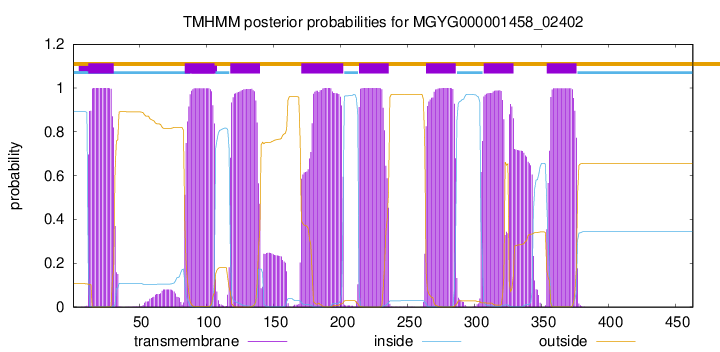
| start | end |
|---|---|
| 12 | 31 |
| 84 | 106 |
| 118 | 140 |
| 171 | 202 |
| 214 | 236 |
| 264 | 286 |
| 307 | 329 |
| 354 | 376 |
