You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001460_02227
You are here: Home > Sequence: MGYG000001460_02227
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Romboutsia dakarensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Romboutsia; Romboutsia dakarensis | |||||||||||
| CAZyme ID | MGYG000001460_02227 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17420; End: 19867 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 329 | 445 | 4.8e-24 | 0.9765625 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033435 | S-layer_Clost | 9.03e-24 | 578 | 807 | 430 | 719 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
| COG4193 | LytD | 9.29e-19 | 303 | 460 | 78 | 241 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam04122 | CW_binding_2 | 1.40e-16 | 546 | 627 | 2 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam01832 | Glucosaminidase | 5.62e-15 | 329 | 392 | 1 | 75 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam04122 | CW_binding_2 | 1.92e-12 | 641 | 722 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CEI73319.1 | 0.0 | 1 | 815 | 1 | 815 |
| CED94675.1 | 1.08e-144 | 3 | 540 | 5 | 557 |
| QSW17879.1 | 8.51e-124 | 130 | 800 | 35 | 751 |
| AOY53018.1 | 1.13e-117 | 52 | 538 | 72 | 581 |
| QQA11780.1 | 1.58e-117 | 52 | 538 | 72 | 581 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J72_A | 1.26e-34 | 538 | 808 | 127 | 428 | ChainA, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630],5J72_B Chain B, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630] |
| 5J6Q_A | 2.85e-33 | 540 | 800 | 283 | 576 | ChainA, Cell wall binding protein cwp8 [Clostridioides difficile 630] |
| 4Q2W_A | 7.46e-23 | 186 | 439 | 62 | 277 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
| 4PI7_A | 4.14e-10 | 303 | 443 | 62 | 210 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.49e-09 | 303 | 443 | 62 | 210 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 3.98e-22 | 186 | 445 | 430 | 651 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| Q02114 | 4.15e-22 | 529 | 807 | 14 | 298 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| P59206 | 4.48e-22 | 186 | 445 | 474 | 695 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| Q8CPQ1 | 1.57e-06 | 307 | 452 | 1165 | 1326 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q5HQB9 | 1.57e-06 | 307 | 452 | 1165 | 1326 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
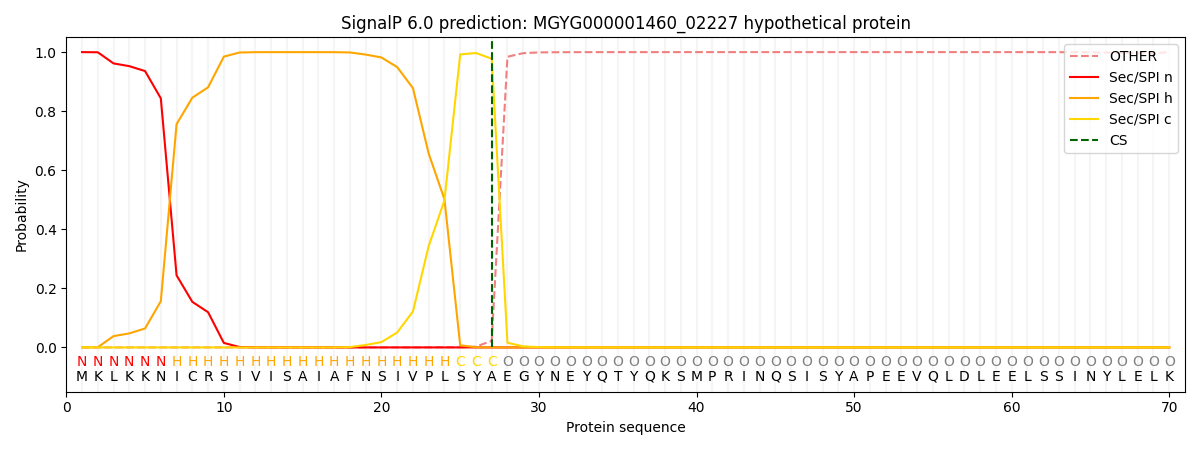
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002584 | 0.995241 | 0.000721 | 0.000543 | 0.000456 | 0.000426 |
