You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001473_02474
You are here: Home > Sequence: MGYG000001473_02474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerosalibacter massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Sporanaerobacteraceae; Anaerosalibacter; Anaerosalibacter massiliensis | |||||||||||
| CAZyme ID | MGYG000001473_02474 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 44195; End: 45418 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 113 | 337 | 6.2e-59 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 8.12e-101 | 52 | 383 | 1 | 319 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.57e-98 | 54 | 375 | 2 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.47e-59 | 62 | 356 | 4 | 298 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 2.26e-20 | 52 | 381 | 45 | 358 | beta-glucosidase BglX. |
| cd01536 | PBP1_ABC_sugar_binding-like | 0.003 | 256 | 361 | 144 | 231 | periplasmic sugar-binding domain of active transport systems that are members of the type 1 periplasmic binding protein (PBP1) superfamily. Periplasmic sugar-binding domain of active transport systems that are members of the type 1 periplasmic binding protein (PBP1) superfamily. The members of this family function as the primary receptors for chemotaxis and transport of many sugar based solutes in bacteria and archaea. The sugar binding domain is also homologous to the ligand-binding domain of eukaryotic receptors such as glutamate receptor (GluR) and DNA-binding transcriptional repressors such as LacI and GalR. Moreover, this periplasmic binding domain, also known as Venus flytrap domain, undergoes transition from an open to a closed conformational state upon the binding of ligands such as lactose, ribose, fructose, xylose, arabinose, galactose/glucose, and other sugars. This family also includes the periplasmic binding domain of autoinducer-2 (AI-2) receptors such as LsrB and LuxP which are highly homologous to periplasmic pentose/hexose sugar-binding proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZY57561.1 | 2.76e-151 | 6 | 395 | 5 | 398 |
| AEE90461.1 | 2.98e-143 | 42 | 401 | 42 | 400 |
| CDI40335.1 | 3.30e-143 | 42 | 401 | 45 | 403 |
| SHD77665.1 | 7.59e-143 | 45 | 403 | 56 | 414 |
| QGU96866.1 | 2.51e-141 | 52 | 401 | 1 | 350 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 1.26e-86 | 54 | 381 | 13 | 342 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3BMX_A | 1.06e-77 | 49 | 381 | 39 | 398 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 3.28e-77 | 49 | 381 | 13 | 372 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 6.47e-77 | 49 | 381 | 43 | 402 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 4ZM6_A | 1.56e-66 | 54 | 391 | 9 | 352 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 5.79e-77 | 49 | 381 | 39 | 398 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| P48823 | 1.09e-56 | 71 | 376 | 51 | 380 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q5QUZ5 | 1.08e-48 | 62 | 350 | 4 | 289 | Beta-hexosaminidase OS=Idiomarina loihiensis (strain ATCC BAA-735 / DSM 15497 / L2-TR) OX=283942 GN=nagZ PE=3 SV=1 |
| Q0AF74 | 2.65e-46 | 62 | 351 | 6 | 296 | Beta-hexosaminidase OS=Nitrosomonas eutropha (strain DSM 101675 / C91 / Nm57) OX=335283 GN=nagZ PE=3 SV=1 |
| Q1H075 | 1.09e-44 | 62 | 339 | 6 | 284 | Beta-hexosaminidase OS=Methylobacillus flagellatus (strain KT / ATCC 51484 / DSM 6875) OX=265072 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
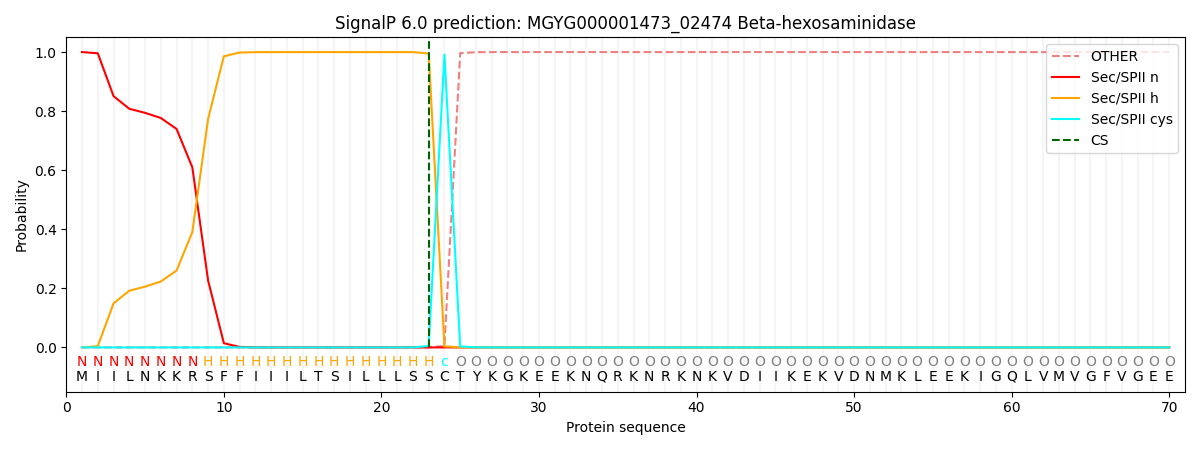
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000002 | 1.000074 | 0.000000 | 0.000000 | 0.000000 |
