You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001480_00023
You are here: Home > Sequence: MGYG000001480_00023
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
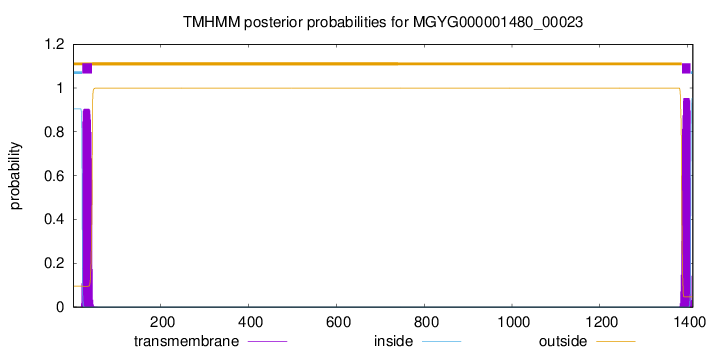
TMHMM annotations
Basic Information help
| Species | Pauljensenia polynesiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia polynesiensis | |||||||||||
| CAZyme ID | MGYG000001480_00023 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10567; End: 14805 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 62 | 602 | 2.4e-174 | 0.9978070175438597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 2.84e-21 | 76 | 415 | 18 | 359 | O-Glycosyl hydrolase family 30. |
| pfam07532 | Big_4 | 1.22e-06 | 778 | 832 | 2 | 56 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam14200 | RicinB_lectin_2 | 3.82e-06 | 659 | 757 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam02057 | Glyco_hydro_59 | 7.53e-06 | 84 | 416 | 9 | 258 | Glycosyl hydrolase family 59. |
| pfam19077 | Big_13 | 5.31e-05 | 1210 | 1260 | 53 | 102 | Bacterial Ig-like domain. Presumed domain found as tandem repeats of high sequence identity in bacterial cell surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ18080.1 | 1.14e-301 | 37 | 1136 | 18 | 1098 |
| QCB92335.1 | 2.97e-301 | 37 | 1134 | 28 | 1103 |
| QVI62156.1 | 7.55e-285 | 60 | 1134 | 40 | 1093 |
| QHC64339.1 | 4.77e-282 | 12 | 1134 | 2 | 1100 |
| QHC56726.1 | 5.46e-281 | 47 | 1134 | 23 | 1089 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 1.71e-10 | 89 | 312 | 36 | 227 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 1.94e-171 | 49 | 1122 | 26 | 1167 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q76FP5 | 3.86e-29 | 61 | 542 | 23 | 422 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
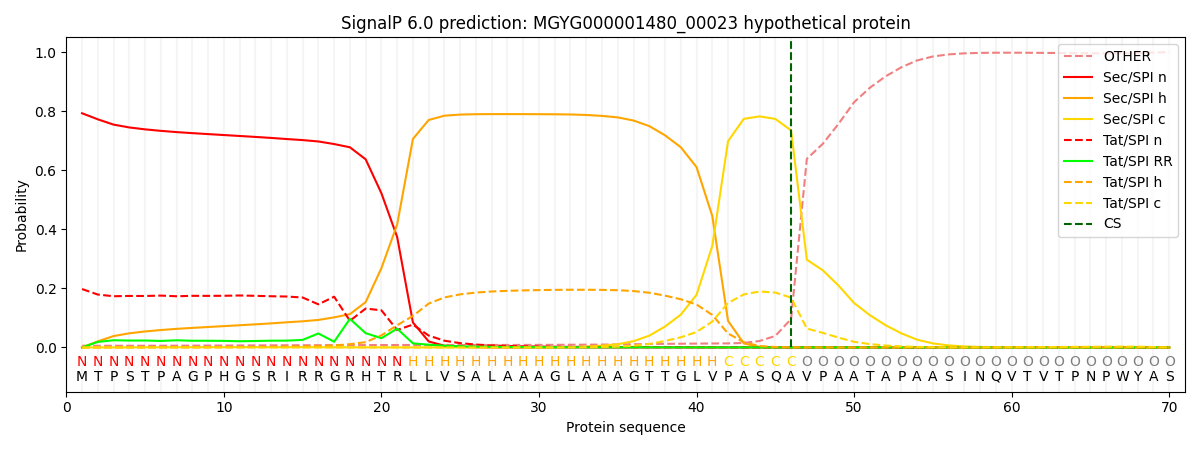
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007432 | 0.785640 | 0.001272 | 0.201796 | 0.003554 | 0.000283 |