You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001480_01057
You are here: Home > Sequence: MGYG000001480_01057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
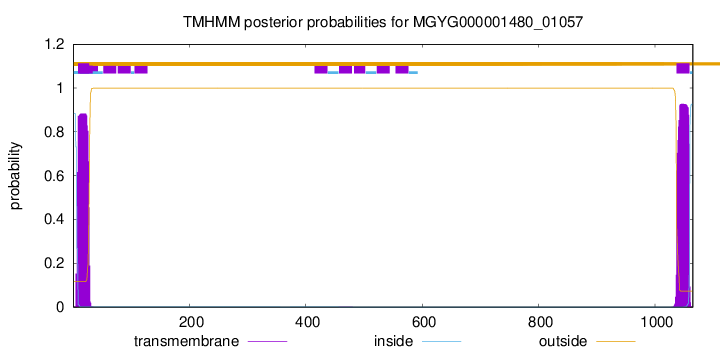
TMHMM annotations
Basic Information help
| Species | Pauljensenia polynesiensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia polynesiensis | |||||||||||
| CAZyme ID | MGYG000001480_01057 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35078; End: 38275 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 723 | 838 | 6.4e-29 | 0.9274193548387096 |
| CBM32 | 591 | 703 | 2.2e-22 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.01e-23 | 723 | 836 | 3 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.49e-14 | 590 | 699 | 2 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| smart00231 | FA58C | 2.03e-11 | 716 | 833 | 8 | 132 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| cd00057 | FA58C | 8.41e-11 | 723 | 826 | 15 | 130 | Substituted updates: Jan 31, 2002 |
| pfam17132 | Glyco_hydro_106 | 7.90e-09 | 532 | 793 | 112 | 369 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFU97473.1 | 4.95e-288 | 2 | 846 | 5 | 840 |
| QKW30838.1 | 3.95e-40 | 590 | 853 | 30 | 293 |
| ACU72398.1 | 4.48e-39 | 591 | 856 | 56 | 329 |
| ACU72589.1 | 2.71e-38 | 594 | 840 | 47 | 297 |
| BAJ26913.1 | 3.73e-38 | 583 | 851 | 31 | 301 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RV9_A | 7.23e-23 | 715 | 841 | 7 | 135 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 7.44e-23 | 715 | 841 | 8 | 136 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 1.38e-22 | 715 | 841 | 8 | 136 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 8.89e-22 | 715 | 841 | 7 | 136 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZZ5_A | 9.15e-22 | 715 | 841 | 8 | 137 | X-raycrystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
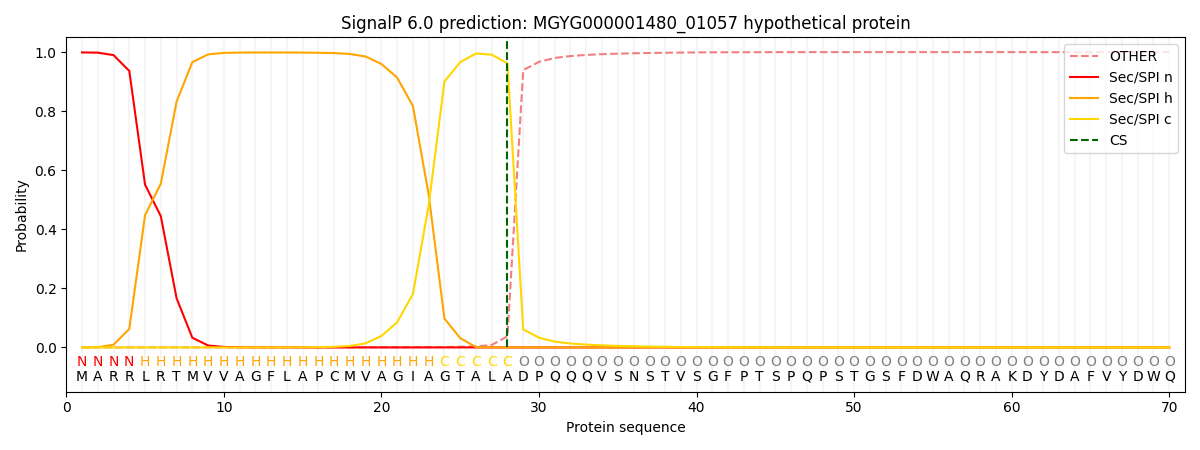
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001255 | 0.996378 | 0.001034 | 0.000797 | 0.000281 | 0.000219 |