You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001481_01831
You are here: Home > Sequence: MGYG000001481_01831
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neobacillus sp000821085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Neobacillus; Neobacillus sp000821085 | |||||||||||
| CAZyme ID | MGYG000001481_01831 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | putative peptidoglycan endopeptidase LytE | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1836588; End: 1837427 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 1.58e-46 | 176 | 276 | 1 | 104 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 1.52e-43 | 26 | 268 | 199 | 471 | invasion associated endopeptidase. |
| COG0791 | Spr | 4.37e-37 | 167 | 276 | 78 | 197 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| PRK10838 | spr | 2.10e-28 | 176 | 279 | 79 | 185 | bifunctional murein DD-endopeptidase/murein LD-carboxypeptidase. |
| PRK06347 | PRK06347 | 4.61e-25 | 23 | 131 | 476 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALC90809.1 | 3.24e-119 | 1 | 279 | 1 | 330 |
| AYA76348.1 | 1.93e-96 | 1 | 277 | 1 | 256 |
| QIZ07902.1 | 2.48e-94 | 1 | 278 | 1 | 256 |
| AND39786.1 | 3.82e-90 | 1 | 277 | 1 | 326 |
| QOK28311.1 | 1.54e-89 | 1 | 277 | 1 | 326 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XCM_A | 1.70e-31 | 88 | 276 | 3 | 228 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 7CFL_A | 7.59e-23 | 164 | 279 | 14 | 138 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4HPE_A | 1.93e-22 | 164 | 275 | 187 | 303 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 2K1G_A | 7.66e-21 | 167 | 279 | 9 | 124 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 4FDY_A | 9.48e-20 | 163 | 279 | 190 | 311 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54421 | 6.02e-70 | 1 | 276 | 1 | 332 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| O07532 | 1.96e-67 | 22 | 276 | 235 | 485 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| O31852 | 4.69e-64 | 29 | 277 | 159 | 412 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| Q01837 | 2.46e-49 | 24 | 274 | 193 | 520 | Probable endopeptidase p60 OS=Listeria ivanovii OX=1638 GN=iap PE=3 SV=1 |
| Q01838 | 9.73e-47 | 26 | 274 | 197 | 519 | Probable endopeptidase p60 OS=Listeria seeligeri OX=1640 GN=iap PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
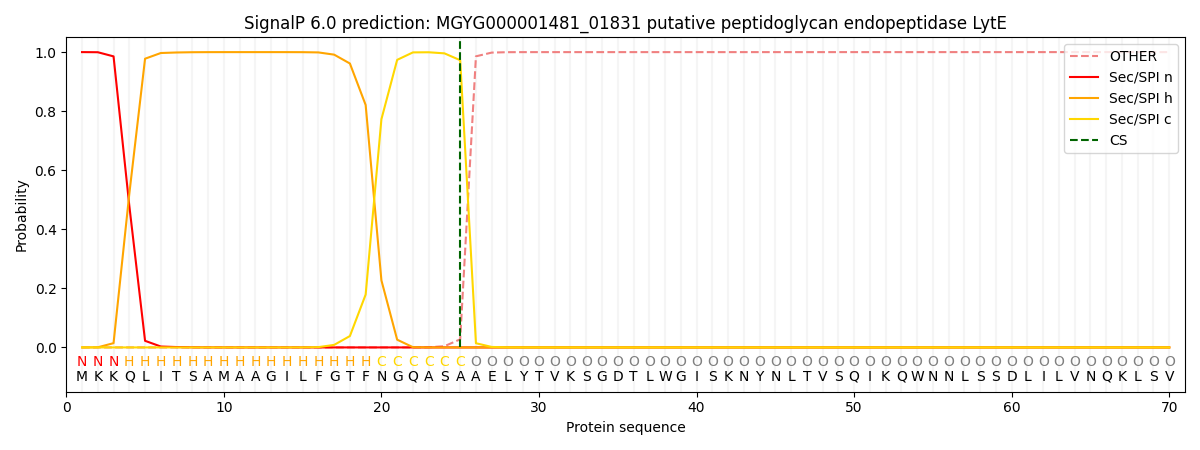
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000220 | 0.999107 | 0.000169 | 0.000177 | 0.000163 | 0.000143 |
