You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001481_03954
You are here: Home > Sequence: MGYG000001481_03954
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Neobacillus sp000821085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Neobacillus; Neobacillus sp000821085 | |||||||||||
| CAZyme ID | MGYG000001481_03954 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3928294; End: 3930537 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 446 | 573 | 1.2e-20 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 8.22e-23 | 393 | 574 | 52 | 228 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 6.49e-13 | 446 | 507 | 1 | 69 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| smart00047 | LYZ2 | 6.58e-09 | 440 | 582 | 7 | 145 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam08239 | SH3_3 | 1.56e-07 | 346 | 398 | 1 | 54 | Bacterial SH3 domain. |
| PRK13914 | PRK13914 | 1.60e-07 | 273 | 405 | 9 | 147 | invasion associated endopeptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZV41185.1 | 6.64e-205 | 1 | 746 | 1 | 752 |
| QIQ32006.1 | 1.33e-199 | 1 | 746 | 1 | 701 |
| QSB49442.1 | 1.55e-195 | 1 | 746 | 1 | 701 |
| QQX25801.1 | 4.57e-195 | 1 | 747 | 1 | 812 |
| QKE74298.1 | 3.84e-193 | 1 | 747 | 1 | 810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.06e-19 | 398 | 574 | 119 | 282 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
| 4PI7_A | 2.24e-09 | 409 | 573 | 52 | 210 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.34e-08 | 409 | 573 | 52 | 210 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 3.97e-08 | 411 | 504 | 62 | 165 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 5WQW_A | 5.43e-06 | 388 | 481 | 20 | 125 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 6.27e-19 | 398 | 574 | 487 | 650 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 6.84e-19 | 398 | 574 | 531 | 694 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| P39848 | 4.99e-11 | 339 | 504 | 619 | 802 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q6GAG0 | 2.59e-09 | 143 | 504 | 853 | 1171 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q6GI31 | 3.41e-09 | 143 | 504 | 860 | 1178 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
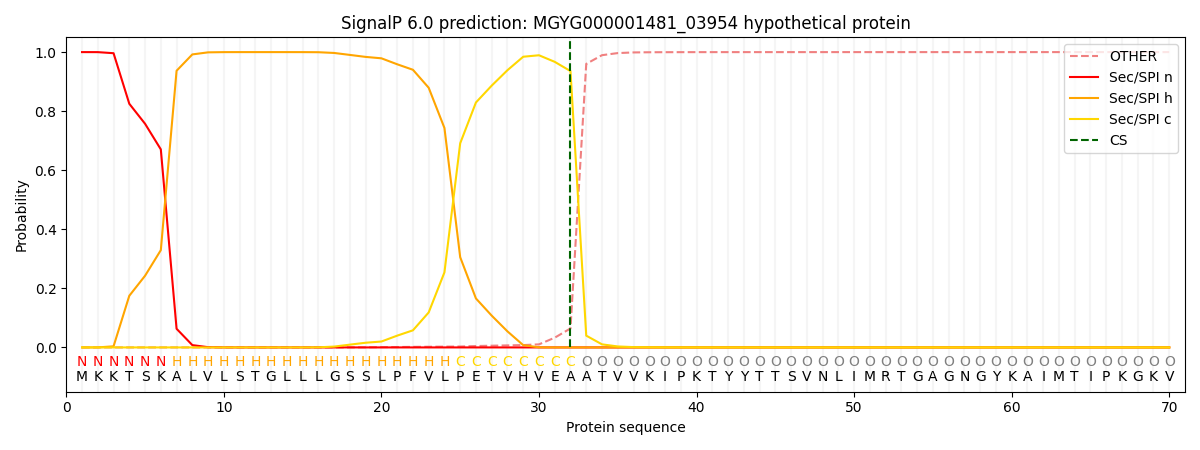
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000273 | 0.999063 | 0.000166 | 0.000185 | 0.000155 | 0.000138 |
