You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001483_00733
You are here: Home > Sequence: MGYG000001483_00733
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Arachnia massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Arachnia; Arachnia massiliensis | |||||||||||
| CAZyme ID | MGYG000001483_00733 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 161887; End: 162714 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06737 | Transglycosylas | 2.03e-29 | 200 | 273 | 1 | 74 | Transglycosylase-like domain. This family of proteins are very likely to act as transglycosylase enzymes related to pfam00062 and pfam01464. These other families are weakly matched by this family, and include the known active site residues. |
| cd13925 | RPF | 6.42e-29 | 202 | 273 | 1 | 70 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
| cd00442 | Lyz-like | 3.72e-04 | 203 | 261 | 1 | 58 | lysozyme-like domains. This family contains several members, including soluble lytic transglycosylases (SLT), goose egg-white lysozymes (GEWL), hen egg-white lysozymes (HEWL), chitinases, bacteriophage lambda lysozymes, endolysins, autolysins, chitosanases, and pesticin. Typical members are involved in the hydrolysis of beta-1,4- linked polysaccharides. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTO37332.1 | 9.65e-160 | 1 | 273 | 1 | 279 |
| AQP51326.1 | 6.47e-43 | 32 | 161 | 1984 | 2117 |
| AQP52241.1 | 8.64e-42 | 34 | 161 | 388 | 520 |
| AQP44644.1 | 1.69e-41 | 31 | 161 | 915 | 1045 |
| QTO38746.1 | 1.64e-40 | 194 | 275 | 301 | 382 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CGE_A | 4.04e-20 | 201 | 271 | 1 | 68 | ChainA, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis],4CGE_B Chain B, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis],4CGE_C Chain C, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis],4CGE_D Chain D, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis],4CGE_E Chain E, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis],4CGE_F Chain F, Resuscitation-promoting Factor Rpfe [Mycobacterium tuberculosis] |
| 4KL7_A | 6.73e-19 | 204 | 271 | 3 | 70 | ChainA, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KL7_B Chain B, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KL7_C Chain C, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KL7_D Chain D, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KPM_A Chain A, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KPM_B Chain B, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KPM_C Chain C, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis],4KPM_D Chain D, Resuscitation-promoting factor RpfB [Mycobacterium tuberculosis] |
| 4EMN_A | 6.91e-19 | 204 | 271 | 4 | 71 | ChainA, Probable resuscitation-promoting factor rpfB [Mycobacterium tuberculosis],4EMN_B Chain B, Probable resuscitation-promoting factor rpfB [Mycobacterium tuberculosis],4EMN_C Chain C, Probable resuscitation-promoting factor rpfB [Mycobacterium tuberculosis],4EMN_D Chain D, Probable resuscitation-promoting factor rpfB [Mycobacterium tuberculosis] |
| 2N5Z_A | 9.92e-19 | 204 | 271 | 9 | 73 | Mycobacteriumtuberculosis: a dynamic view of the resuscitation promoting factor RpfC catalytic domain [Mycobacterium tuberculosis H37Rv] |
| 4OW1_A | 1.30e-18 | 204 | 271 | 6 | 70 | ChainA, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_B Chain B, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_E Chain E, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_S Chain S, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_T Chain T, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_U Chain U, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_W Chain W, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis],4OW1_X Chain X, Resuscitation-promoting factor RpfC [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6M6W5 | 2.86e-22 | 204 | 271 | 41 | 108 | Resuscitation-promoting factor Rpf1 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=rpf1 PE=3 SV=1 |
| O53177 | 2.66e-19 | 176 | 271 | 79 | 165 | Resuscitation-promoting factor RpfE OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=rpfE PE=1 SV=1 |
| O86308 | 4.11e-18 | 162 | 271 | 5 | 111 | Resuscitation-promoting factor Rpf OS=Micrococcus luteus OX=1270 GN=rpf PE=1 SV=2 |
| P9WG30 | 1.37e-17 | 194 | 271 | 34 | 111 | Resuscitation-promoting factor RpfA OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=rpfA PE=3 SV=1 |
| P9WG31 | 1.37e-17 | 194 | 271 | 34 | 111 | Resuscitation-promoting factor RpfA OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=rpfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
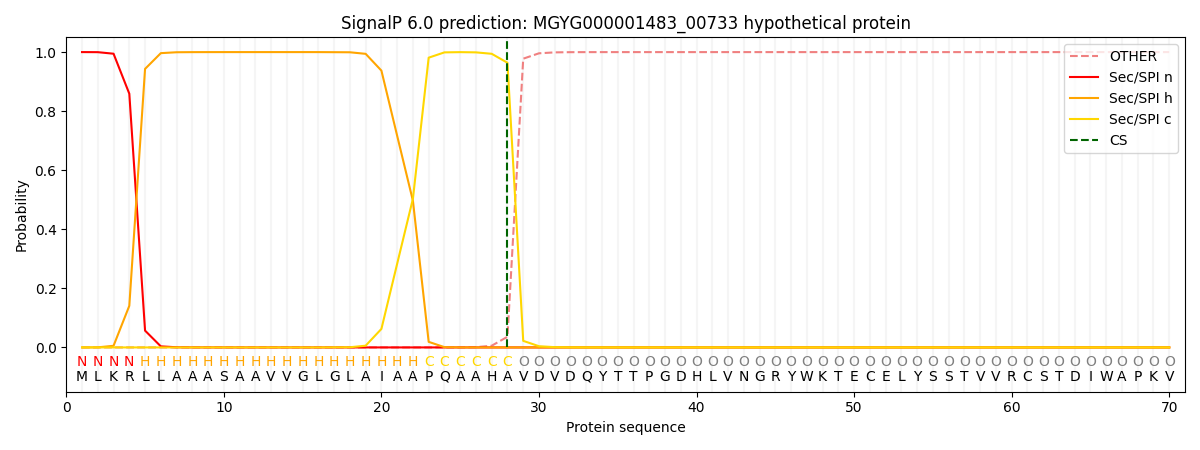
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000210 | 0.999136 | 0.000157 | 0.000186 | 0.000157 | 0.000139 |
