You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001483_03033
You are here: Home > Sequence: MGYG000001483_03033
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
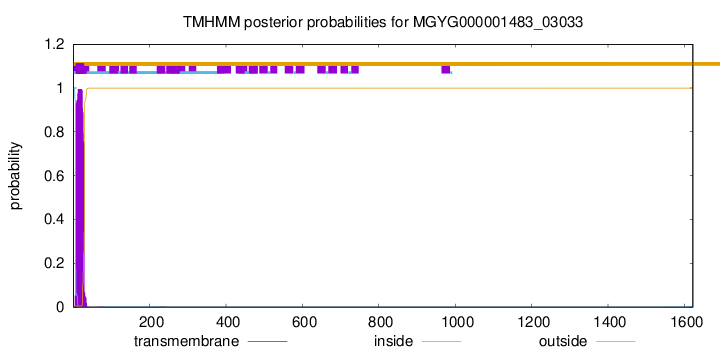
TMHMM annotations
Basic Information help
| Species | Arachnia massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Arachnia; Arachnia massiliensis | |||||||||||
| CAZyme ID | MGYG000001483_03033 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 929206; End: 934074 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 36 | 586 | 8e-159 | 0.9543795620437956 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 9.25e-12 | 1180 | 1327 | 16 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam12708 | Pectate_lyase_3 | 3.79e-08 | 37 | 79 | 2 | 44 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam05345 | He_PIG | 1.40e-06 | 1375 | 1456 | 1 | 94 | Putative Ig domain. This alignment represents the conserved core region of ~90 residue repeat found in several haemagglutinins and other cell surface proteins. Sequence similarities to (pfam02494) and (pfam00801) suggest an Ig-like fold (personal obs:C. Yeats). So this family may be similar in function to the (pfam02639) and (pfam02638) domains. This domain is also found in the WisP family of proteins of Tropheryma whipplei. |
| pfam13229 | Beta_helix | 2.48e-05 | 456 | 581 | 12 | 134 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| PLN02188 | PLN02188 | 2.63e-04 | 26 | 79 | 26 | 79 | polygalacturonase/glycoside hydrolase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH68773.1 | 1.78e-208 | 31 | 1063 | 78 | 1010 |
| ANJ28196.1 | 2.21e-198 | 36 | 1042 | 68 | 985 |
| CAJ90659.1 | 7.57e-177 | 22 | 701 | 20 | 704 |
| AQW50555.1 | 1.51e-173 | 35 | 716 | 67 | 761 |
| ASQ94414.1 | 3.36e-171 | 35 | 716 | 13 | 707 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 6.20e-47 | 22 | 581 | 13 | 566 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 2.71e-46 | 22 | 581 | 13 | 566 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
| 1W8N_A | 1.30e-12 | 1098 | 1247 | 402 | 540 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 2BZD_A | 1.30e-12 | 1098 | 1247 | 402 | 540 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
| 2BER_A | 1.30e-12 | 1098 | 1247 | 402 | 540 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1V8K7 | 1.51e-177 | 22 | 701 | 20 | 704 | Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
| Q826C5 | 2.41e-165 | 25 | 644 | 29 | 624 | Alpha-1,3-galactosidase A OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=glaA PE=3 SV=1 |
| Q64MU6 | 3.42e-84 | 39 | 586 | 28 | 555 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
| Q5L7M8 | 1.57e-83 | 39 | 586 | 28 | 555 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaA PE=3 SV=1 |
| Q8A2Z5 | 1.66e-66 | 54 | 586 | 29 | 537 | Alpha-1,3-galactosidase A OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
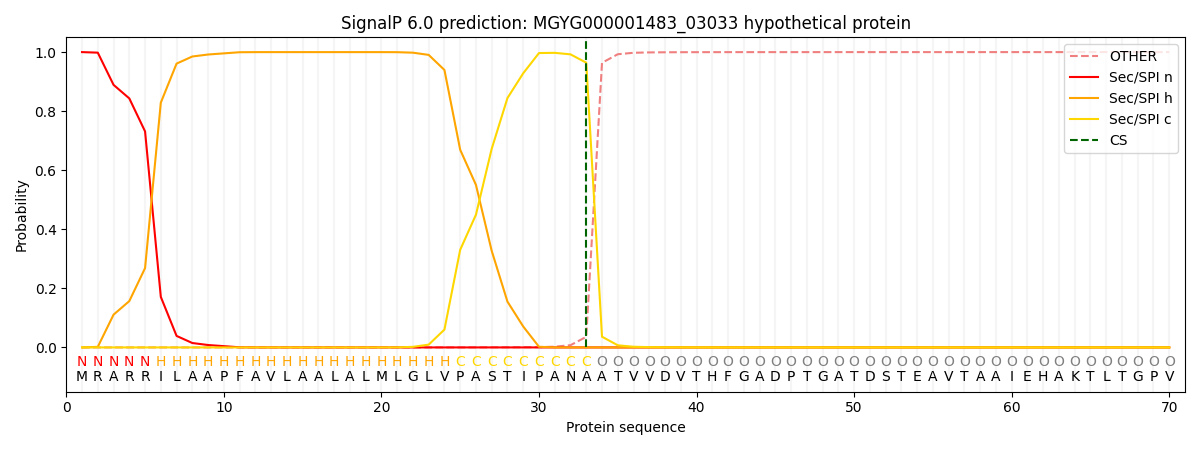
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000260 | 0.999084 | 0.000149 | 0.000198 | 0.000150 | 0.000135 |