You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001485_02030
You are here: Home > Sequence: MGYG000001485_02030
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
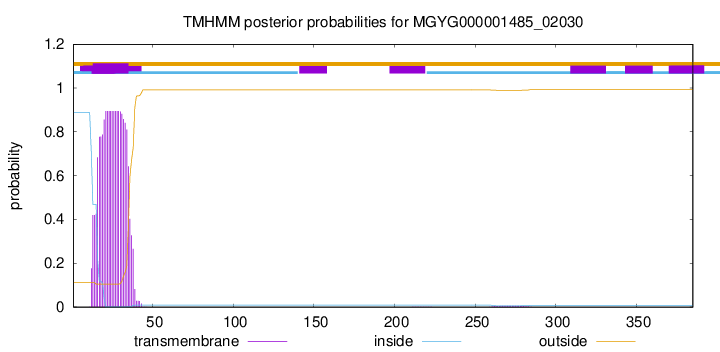
TMHMM annotations
Basic Information help
| Species | Pseudomonas_E massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_E; Pseudomonas_E massiliensis | |||||||||||
| CAZyme ID | MGYG000001485_02030 | |||||||||||
| CAZy Family | PL5 | |||||||||||
| CAZyme Description | Alginate lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 886399; End: 887556 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL5 | 34 | 349 | 4.2e-181 | 0.9936708860759493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK00325 | algL | 0.0 | 14 | 372 | 2 | 359 | polysaccharide lyase. |
| cd00244 | AlgLyase | 0.0 | 30 | 368 | 1 | 339 | Alginate Lyase A1-III; enzymatically depolymerizes alginate, a complex copolymer of beta-D-mannuronate and alpha-L-guluronate, by cleaving the beta-(1,4) glycosidic bond. |
| pfam05426 | Alginate_lyase | 7.39e-64 | 69 | 310 | 1 | 274 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFS53362.1 | 3.45e-211 | 14 | 378 | 7 | 373 |
| AZD27771.1 | 3.45e-211 | 14 | 378 | 7 | 373 |
| QIT21105.1 | 6.95e-211 | 14 | 378 | 7 | 373 |
| BBN52925.1 | 6.95e-211 | 14 | 378 | 7 | 373 |
| AZD46469.1 | 6.95e-211 | 14 | 378 | 7 | 373 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OZV_A | 1.11e-173 | 34 | 368 | 1 | 334 | CrystalStructure of the periplasmic alginate lyase AlgL [Pseudomonas aeruginosa PAO1] |
| 4OZW_A | 3.68e-172 | 34 | 368 | 1 | 334 | CrystalStructure of the periplasmic alginate lyase AlgL H202A mutant [Pseudomonas aeruginosa PAO1] |
| 7FHZ_A | 1.14e-46 | 59 | 365 | 1 | 297 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FHU_A | 1.36e-46 | 59 | 365 | 1 | 297 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHU_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI0_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI1_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FI2_A | 3.97e-45 | 59 | 365 | 1 | 297 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9ZNB7 | 1.03e-206 | 14 | 378 | 7 | 373 | Alginate lyase OS=Cobetia marina OX=28258 GN=algL PE=3 SV=1 |
| Q3KHR0 | 2.96e-206 | 14 | 378 | 7 | 373 | Alginate lyase OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=algL PE=3 SV=1 |
| Q4KHY5 | 4.37e-205 | 14 | 375 | 7 | 370 | Alginate lyase OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=algL PE=3 SV=1 |
| P59786 | 4.48e-203 | 14 | 378 | 6 | 372 | Alginate lyase OS=Pseudomonas fluorescens OX=294 GN=algL PE=3 SV=1 |
| C3KDZ3 | 4.48e-203 | 14 | 378 | 6 | 372 | Alginate lyase OS=Pseudomonas fluorescens (strain SBW25) OX=216595 GN=algL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
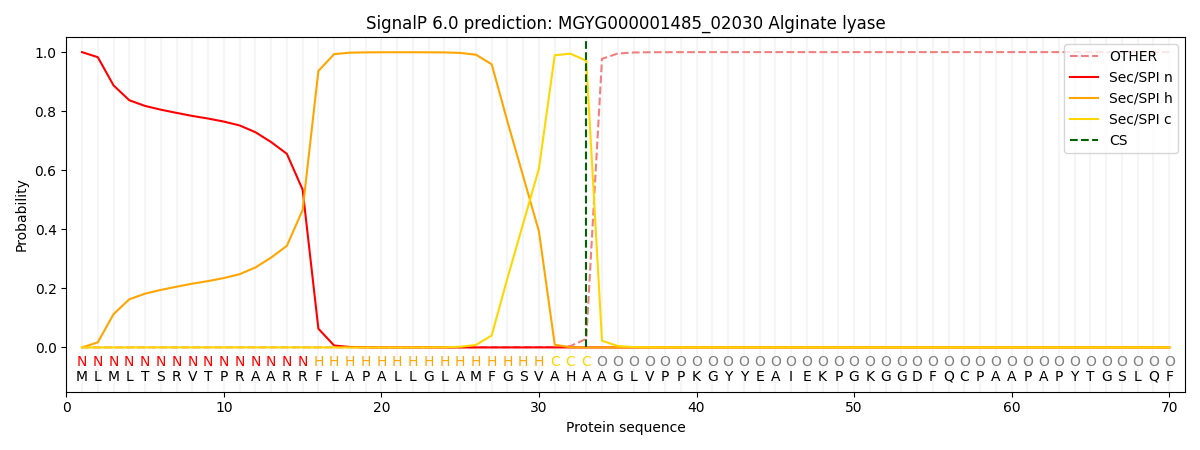
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000776 | 0.998170 | 0.000250 | 0.000345 | 0.000220 | 0.000184 |