You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001486_00084
Basic Information
help
| Species |
Mobilicoccus massiliensis
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Dermatophilaceae; Mobilicoccus; Mobilicoccus massiliensis
|
| CAZyme ID |
MGYG000001486_00084
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001486 |
3841955 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 62610;
End: 64751
Strand: +
|
No EC number prediction in MGYG000001486_00084.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
118 |
505 |
8.5e-35 |
0.6907407407407408 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
3.96e-14 |
86 |
511 |
33 |
435 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
4.49e-14 |
119 |
237 |
1 |
119 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P37483
|
8.35e-51 |
63 |
679 |
12 |
654 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
|
O34575
|
3.10e-48 |
88 |
676 |
34 |
664 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.973694
|
0.025299
|
0.000450
|
0.000228
|
0.000118
|
0.000215
|
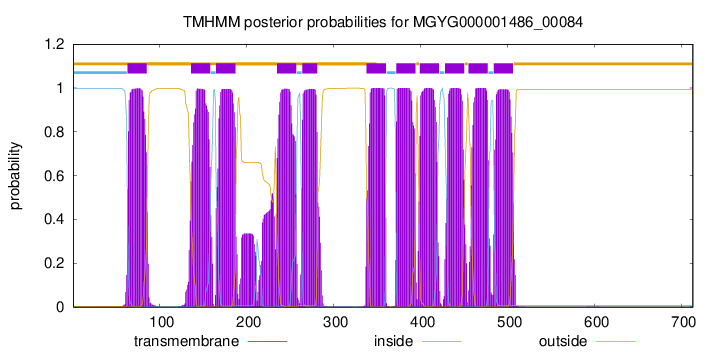
| start |
end |
| 63 |
85 |
| 136 |
158 |
| 165 |
187 |
| 235 |
257 |
| 264 |
281 |
| 338 |
360 |
| 372 |
394 |
| 399 |
421 |
| 428 |
450 |
| 455 |
477 |
| 484 |
506 |


