You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001486_00793
You are here: Home > Sequence: MGYG000001486_00793
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
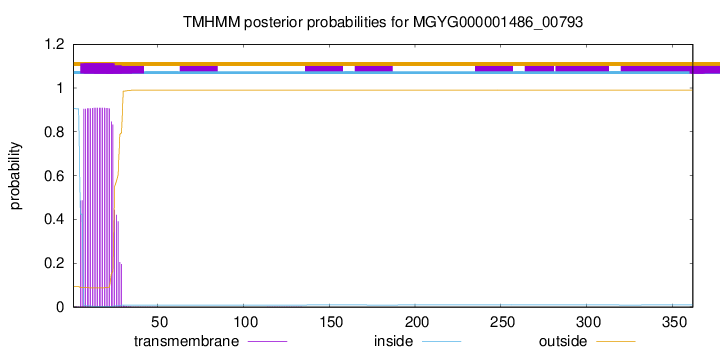
TMHMM annotations
Basic Information help
| Species | Mobilicoccus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Dermatophilaceae; Mobilicoccus; Mobilicoccus massiliensis | |||||||||||
| CAZyme ID | MGYG000001486_00793 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 286189; End: 287277 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 146 | 269 | 3.6e-16 | 0.35973597359735976 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SNV16686.1 | 4.81e-125 | 64 | 362 | 60 | 358 |
| SDS94672.1 | 3.61e-107 | 55 | 362 | 53 | 359 |
| SDS59482.1 | 1.74e-90 | 55 | 362 | 92 | 399 |
| ART72432.1 | 4.24e-51 | 68 | 362 | 22 | 304 |
| CCG01385.1 | 1.98e-43 | 63 | 362 | 155 | 445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVT_A | 1.90e-06 | 170 | 310 | 168 | 296 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 2X2Y_A | 1.90e-06 | 170 | 310 | 168 | 296 | Cellulomonasfimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi],2X2Y_B Cellulomonas fimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi] |
| 3TP4_A | 4.43e-06 | 170 | 310 | 168 | 296 | CrystalStructure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct],3TP4_B Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
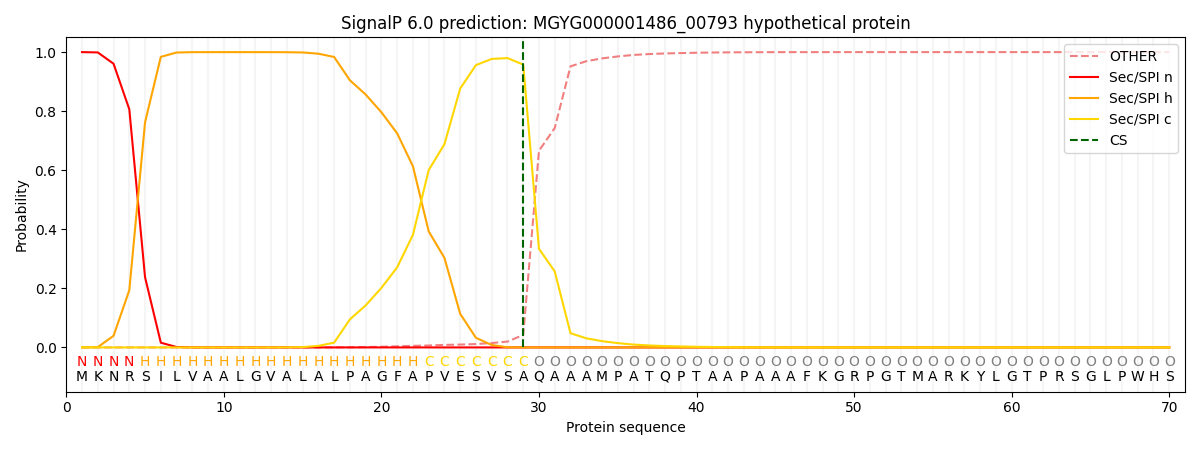
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000380 | 0.998745 | 0.000258 | 0.000241 | 0.000196 | 0.000153 |