You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001486_02056
You are here: Home > Sequence: MGYG000001486_02056
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mobilicoccus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Dermatophilaceae; Mobilicoccus; Mobilicoccus massiliensis | |||||||||||
| CAZyme ID | MGYG000001486_02056 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 102603; End: 104204 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 139 | 274 | 2.9e-20 | 0.7851851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13399 | Slt35-like | 3.91e-23 | 136 | 276 | 1 | 108 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| COG2951 | MltB | 6.51e-21 | 112 | 286 | 106 | 287 | Membrane-bound lytic murein transglycosylase B [Cell wall/membrane/envelope biogenesis]. |
| pfam13406 | SLT_2 | 2.35e-07 | 187 | 240 | 139 | 196 | Transglycosylase SLT domain. This family is related to the SLT domain pfam01464. |
| PHA03247 | PHA03247 | 9.69e-06 | 272 | 497 | 2634 | 2844 | large tegument protein UL36; Provisional |
| PHA03247 | PHA03247 | 6.50e-05 | 298 | 496 | 2685 | 2879 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIZ38977.1 | 1.51e-56 | 111 | 285 | 99 | 273 |
| BAH54290.1 | 1.84e-55 | 68 | 285 | 77 | 286 |
| QNG22248.1 | 2.09e-55 | 42 | 283 | 25 | 273 |
| QNG18082.1 | 2.09e-55 | 42 | 283 | 25 | 273 |
| BCN58127.1 | 2.43e-55 | 80 | 285 | 19 | 227 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
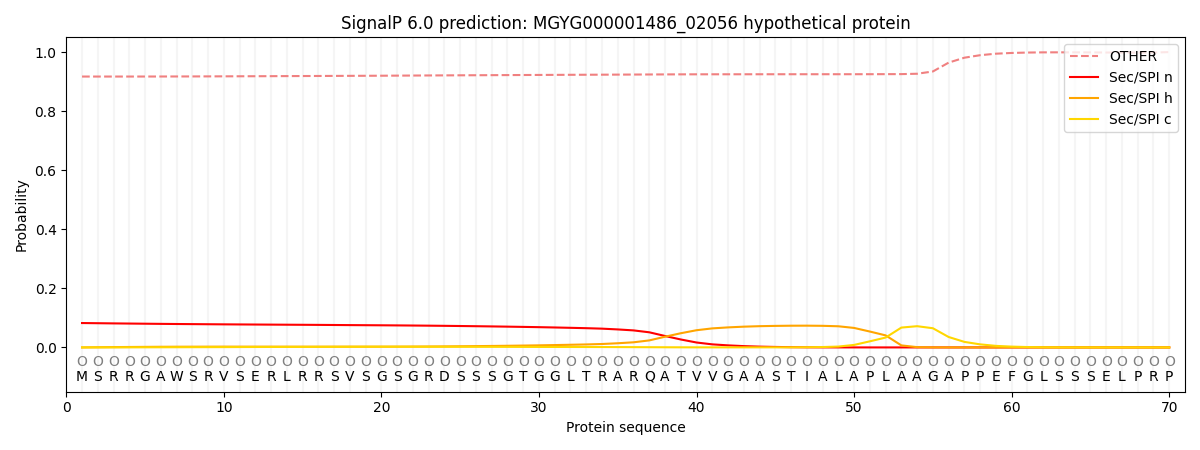
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.921815 | 0.076811 | 0.000446 | 0.000461 | 0.000212 | 0.000247 |
